1. Introduction
Proteins are commonly created in heterologous structures because it is tough to acquire high-quality products from herbal sources. The efficient expression and purification of recombinant proteins continue to be fundamental problems in biotechnology. Several methods have been used to simplify the purification procedure, including the method of affinity [
1,
2]. Because figuring out the structure of the relationship between a protein and a ligand is important for biochemistry and finding new drugs [
3,
4], protein purification is an important method for both academia and industry [
5,
6].
The isolation, separation, and purification of diverse proteins, peptides, and specific molecules are employed in nearly all branches of bioscience and biotechnology. For this reason, the advancement of separation knowledge and technology is an important characteristic of bio-oriented research and development. “New separation techniques are needed that can treat dilute solutions or solutions with only small amounts of target molecules in the presence of a large number of other compounds, even in the particulate matter” [
7].
The improvement and accuracy of proteomics are inseparable from separation and identification development. Natural proteins are often mixtures, and many critical proteins are poor in biomaterials. Therefore, the isolation of proteins without affecting their shape, composition, and activity has continually been an issue in proteomics research [
8]. With the improvements in development, many new technologies have been implemented for protein separation, such as the multi-column plate adapter (MCPA) [
9], the cell-surface display primarily based on the SUMO-Ulp1 system [
10], the aqueous two-phase system (ATPS) [
11], chromatography [
11,
12], magnetic separation [
7,
13], etc. According to the protein residence, it is becoming more common to cleave proteins by combining a couple of methods that can better hold the function and shape of the proteins and obtain better resolution.
This paper mainly reviews and discusses one of the core components of proteomics—the latest advancements in separation technology for protein components—focusing on five different methods.
2. Multi-Column Plate Adapter (MCPA) System
Many studies on protein structure and function require multiple protein purification techniques to develop protein purification methods. The case of the purification of therapeutic proteins, such as antibodies, is referred to. Since then, technologies have been developed to meet the demand. However, higher protein yields and purity in one step come with a higher price, which is usually not feasible for small-scale research [
14]. Immobilized metal affinity chromatography (IMAC), automated fast protein liquid chromatography (FPLC), and protein maker are the instruments that are used now.
Some researchers believe a cheaper protein purification method with higher purity and a shorter duration can be developed with a multi-well collection plate and simple gravity. This is the main idea behind the multi-column plate adapter (MCPA) system. The system can be set up in different ways and with various conditions, such as other types of resin, buffer systems, wash buffer imidazole concentration, and lysate load volume [
15].
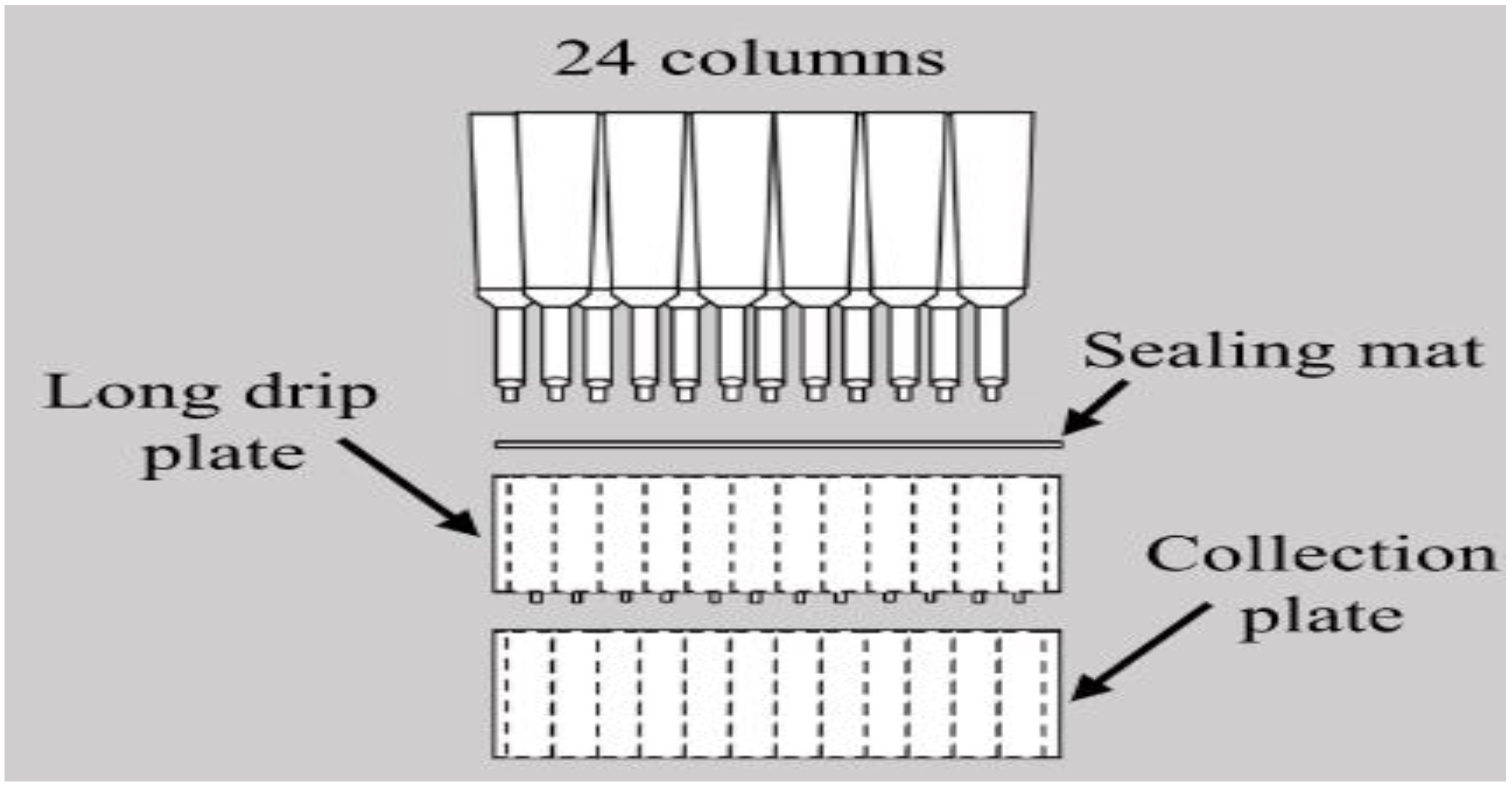
The MCPA system consists of a series of long-drip filter plates with a sealing mat and other conventional laboratory supplies, as in
Figure 1 and
Figure 2. The system can be reused repeatedly for protein purification after cleaning. The system uses affinity chromatography as a protein purification technique. Some researchers used ion-exchange column chromatography with the method [
16].
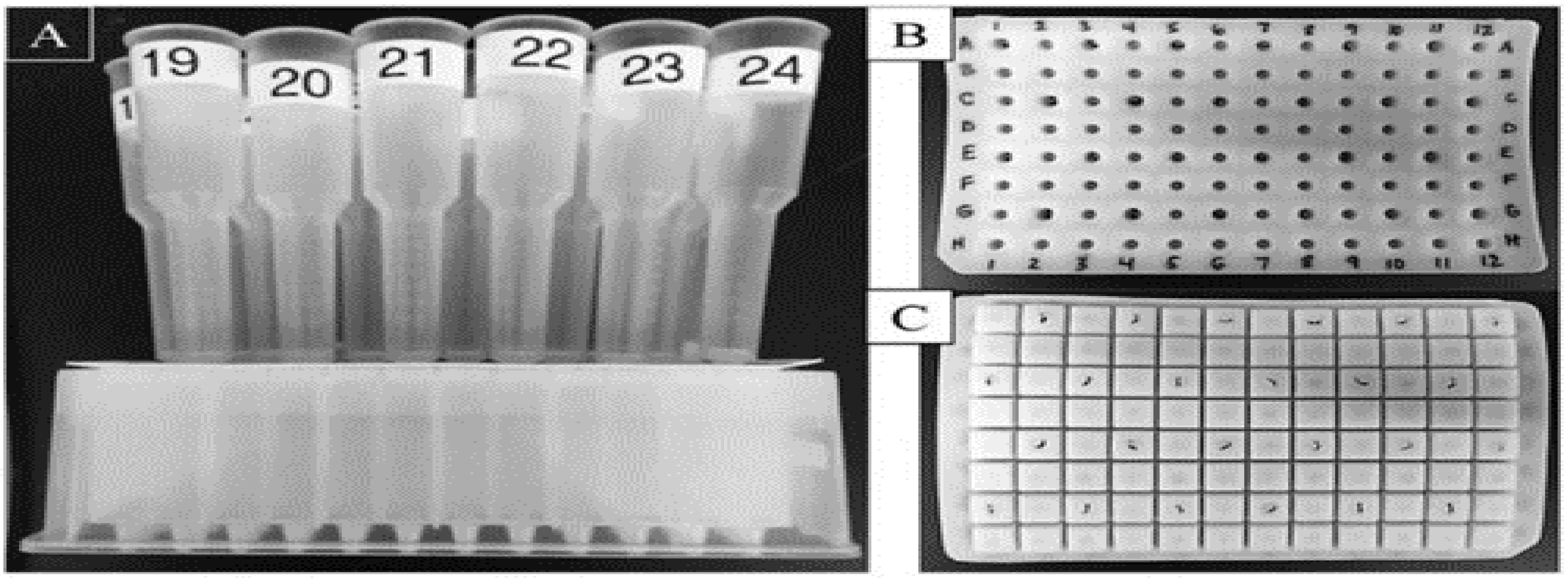
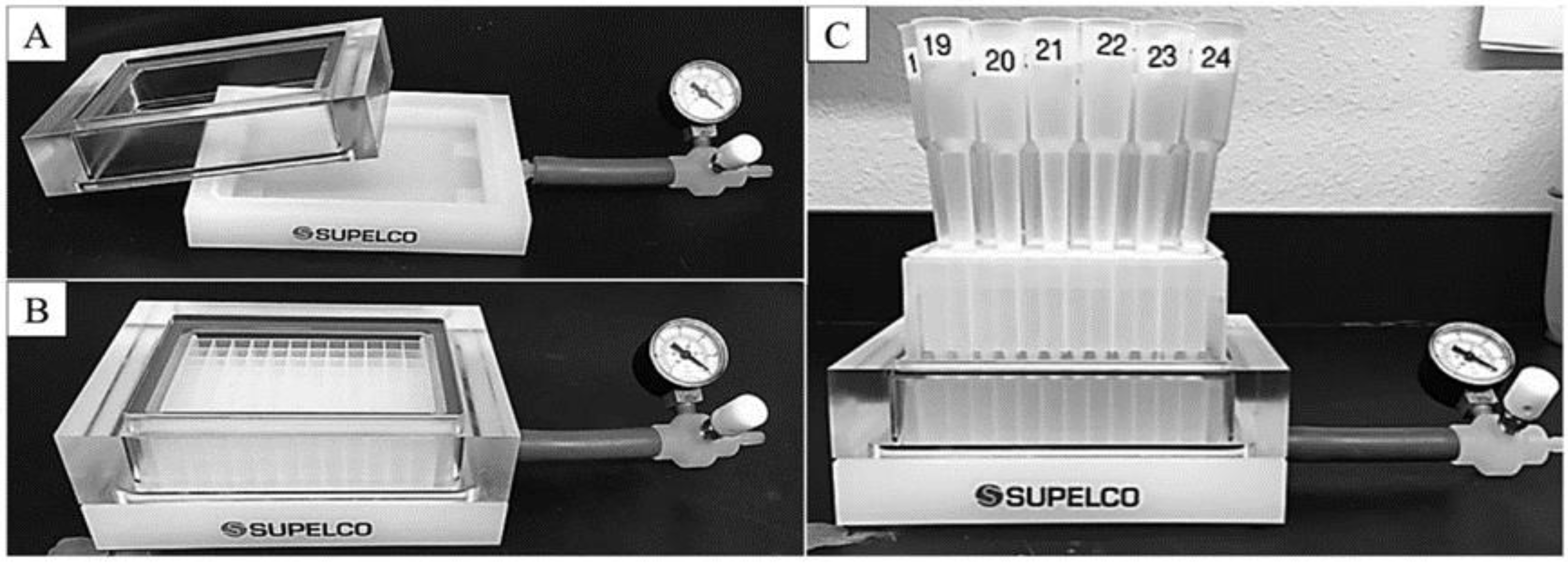
A study of an MCPA system with affinity chromatography and without vacuum that cost only USD 45 worked well to purify proteins, as summarised in
Figure 3. In
Figure 4, vacuum manifolds greatly assisted the purification process by completing it in less time than gravity.
The study successfully purified yeast AbpSH3 mutants using nickel (Ni) resins under denaturing and native purification conditions. This was indicated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Under native purification conditions, the samples with common contaminants at 25 kDa found under denaturing conditions [
9] were purified.
The protein purification method has been scaled up from one column per protein for 24 different samples to 12 columns per protein for two different samples. The samples were highly pure with the same contaminants as those obtained using small-scale protein purification. Reference [
9] found that the MCPA system for protein purification also made enough protein for biophysical analysis, such as circular dichroism (CD) spectroscopy.
3. Cell-Surface Display Based on SUMO–Ulp1 System
The cell-surface display method provides the opportunity to target proteins on the surface of microbial cells, and fusion with an anchoring motif completes the cell-surface display. This technique is known as a practical method for numerous applications, including in the development of vaccines, environmental bio-adsorbents, and whole-cell biocatalysts [
17]. Recently, this technique was suggested as able to simplify protein purification. Reference [
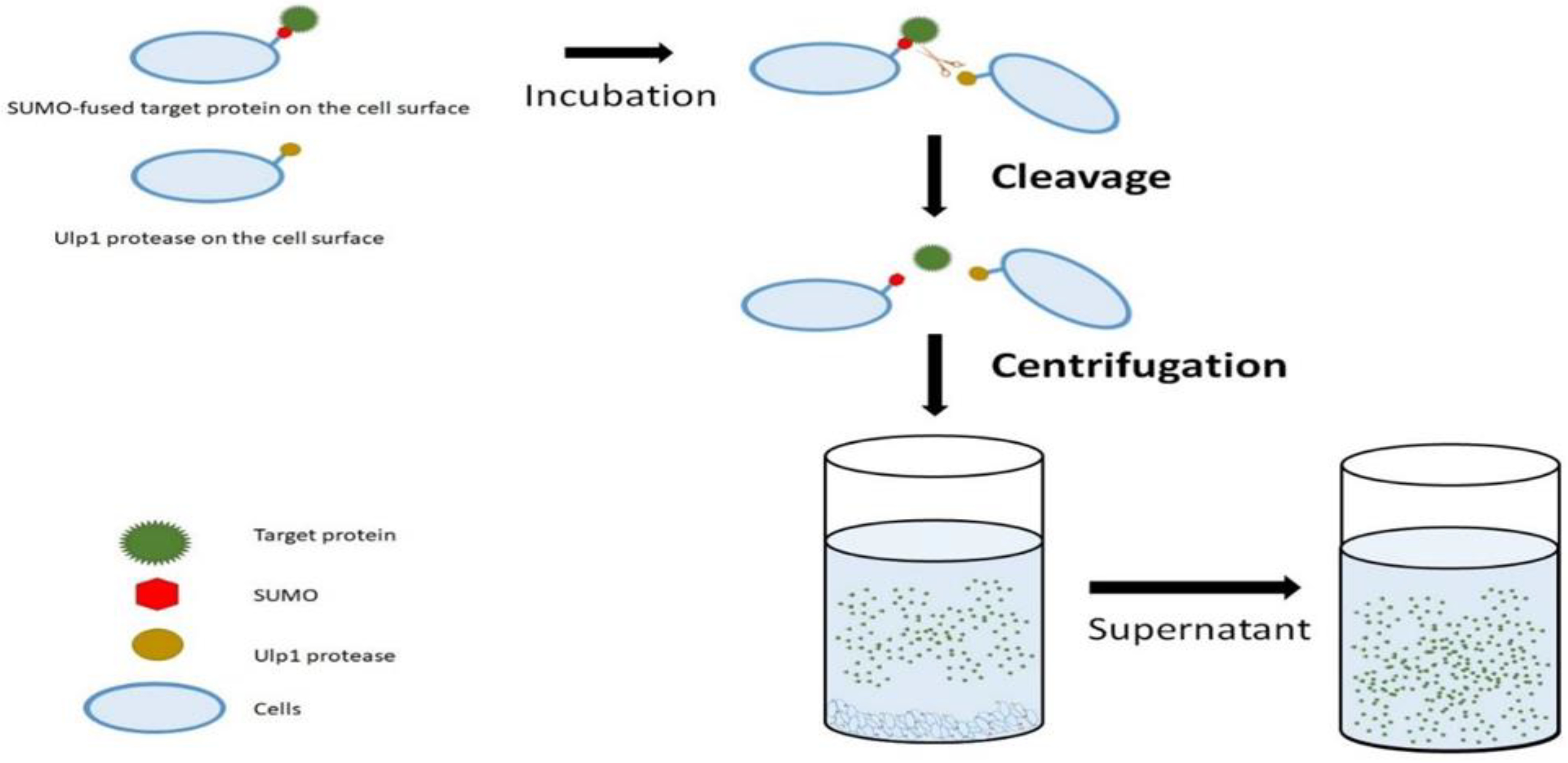
10] reported the application of cell-surface display in protein purification based on the cleavage of a SUMO-fused target protein by Ulp1 protease that was revealed on the surface of Escherichia coli cells, as described above (
Figure 5). SUMO, a ubiquitin-like protein, has been used to improve target protein stability and solubility through N-terminal fusion [
18,
19]. Ulp1 protease can cut the SUMO tag, which breaks the SUMO tertiary configuration and makes a native target protein with no extra amino acids [
20].
The effectiveness of the SUMO-Ulp1 structure in protein purification can be archived using two different vectors: (i) the expression of SUMO-fused target protein on cell surfaces and (ii) the expression of Ulp1 protease on cell surfaces [
10]. In this system, the N-terminal of a SUMO-fused target protein will be cleaved by the surface-displayed Ulp1 protease. This leads to the release of native target proteins in the buffer solution. Surface-displayed SUMO and Ulp1 protease can be removed together with the cells by centrifugation. As a result, the target protein can be collected in the form of a supernatant after centrifugation and further improved in its purity by simple ultrafiltration. For example, it has been reported that the purity of the target protein obtained was recorded at more than 80% using the SUMO-Ulp1 system. The target protein purity was improved by more than 90% using simple ultrafiltration [
10]. Overall, this technique is a simple way to purify the target protein because it only needs the cleavage and centrifugation steps.
4. Cell-Surface Display Based on SUMO-Ulp1 System
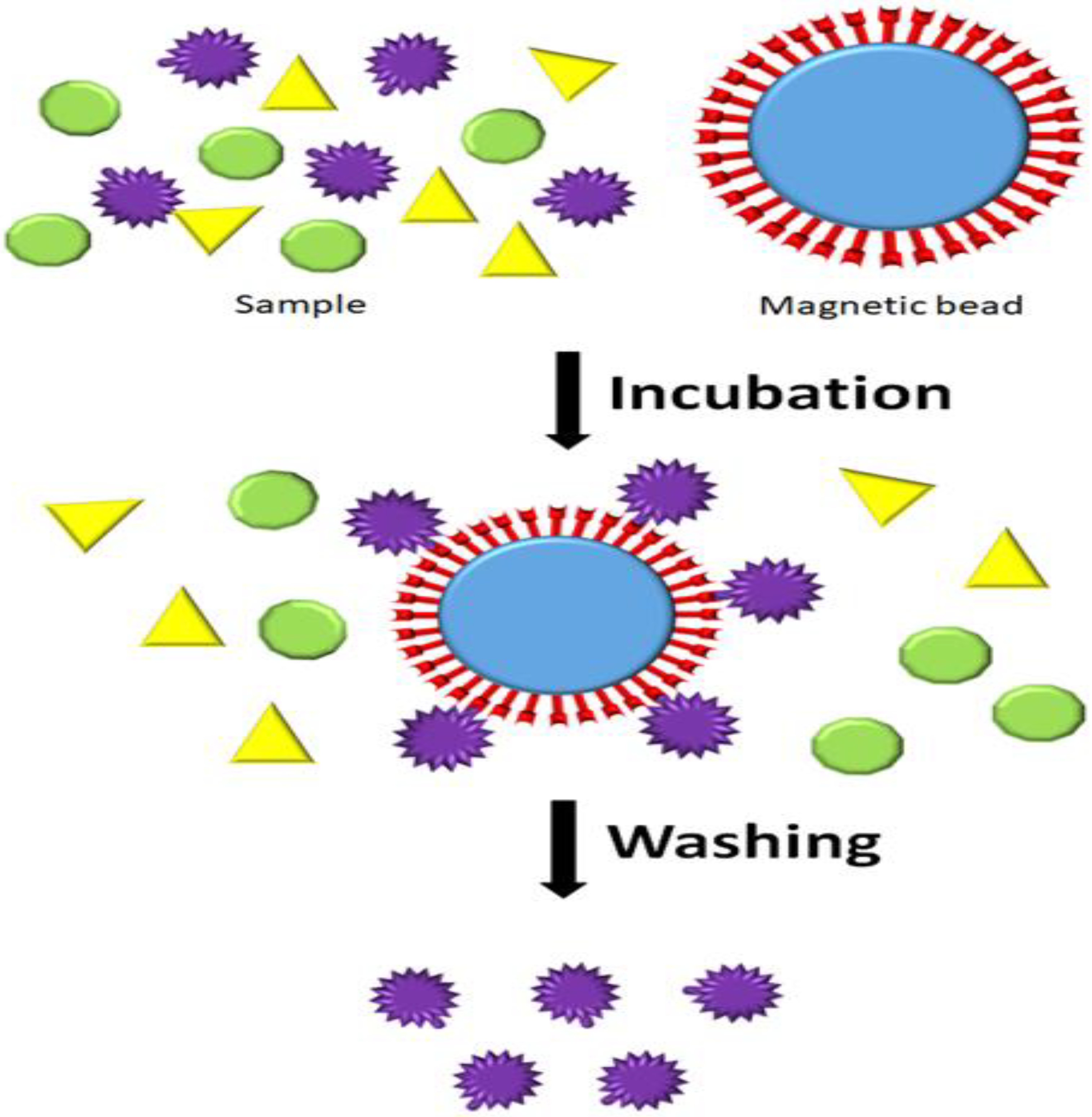
In protein purification, the magnetic separation technique offers a simple approach for obtaining a high purity level of the target protein by applying magnetic beads. Magnetic beads are magnetic carriers and affinity ligands [
7]. The target protein is bound explicitly to the specific ligands on the magnetic beads’ surface, creating a magnetic complex during incubation (
Figure 6). Surprisingly, a magnetic separator can easily and quickly extract the target protein from the magnetic complex. After the removal step, the washing step can be used to further separate the target protein from the other things that are not what they should be.
Magnetic separation techniques, in addition to their cost-effectiveness, offer several advantages, including high yield and binding capacity, sensitivity, as well as reproducibility [
21,
22]. Using this technique, the separation procedure can be conducted openly in primary samples, which shortens the total purification period. Furthermore, this technique could be helpful for large-scale operations because of its strength and efficiency. As a part of the technique described above, magnetic separation also offers a very gentle process for the purification of the target protein. Using a magnetic separation method to purify large protein complexes has been shown to obtain more stable proteins than the traditional column chromatography method [
23].
5. Aqueous Two-Phase System (ATPS)
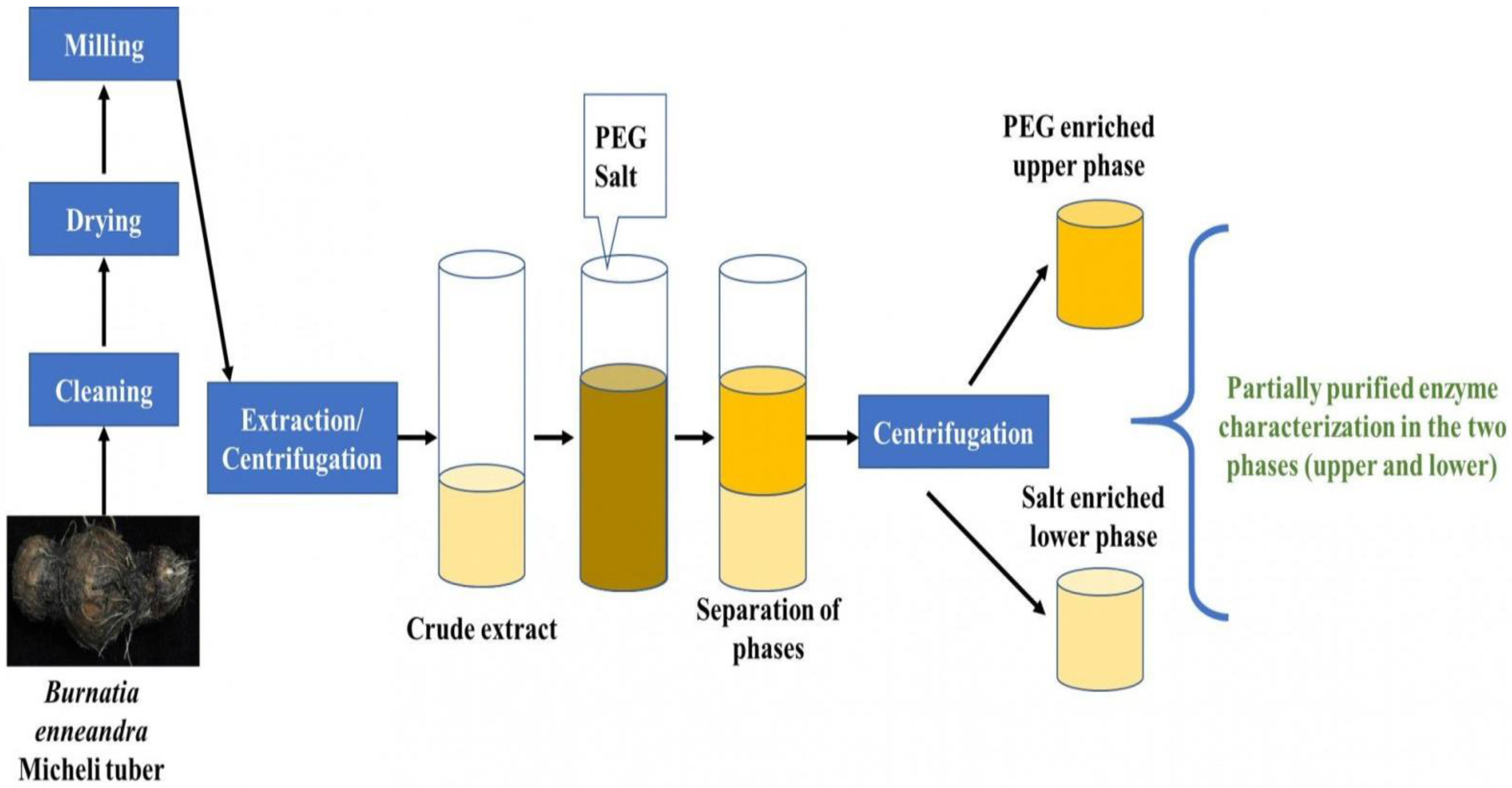
The “aqueous two-phase system (ATPS) is a liquid-liquid separation technique that has shown great potential for the extraction, recovery, and purification of a great variety of biological compounds” [
24]. It is a technique commonly used to separate and purify enzymes and proteins. The advantages of ATPS in protein purification include the simplicity and speed of the separation with minimal denaturation of the enzymes [
25]. The most important part of both phases is water (80 to 90%), and most polymers stabilise protein structure [
26]. The ATPS process is shown in
Figure 7.
There are several types of ATPS, such as polymeric aqueous two-phase systems (ATPS-P) and micellar aqueous two-phase systems (ATPS-M). Additionally, there are reverse aqueous two-phase micellar systems (ATPS-RM) and ionic liquid-based aqueous two-phase systems [
27].
6. Types of ATPS
6.1. Polymeric Aqueous Two-Phase System
The “polymeric aqueous two-phase system (ATPS-P) is a liquid-liquid purification technique using polymers and salt solutions or polymer/polymer solution mixtures” [
29]. Two water-soluble solutes separate into two immiscible aqueous-rich phases based on polymer–polymer, polymer–salt, or salt–salt solute combinations [
29]. The advantage of this structure is that it is simply due to the combination of polymer and salt, even though the price of polymers can be high compared to the cost of salt.
6.2. Micellar Aqueous Two-Phase System
“Micellar aqueous two-phase systems (ATPS-M) are formed by surfactant solution. Surfactants are amphiphilic molecules containing a hydrophilic (the head) and a hydrophobic (the tail). The surfactant head can be charged either anionic or cationic, dipolar (zwitterionic), or non-charged (non-ionic)” [
30]. Surfactants form micelle solutions, and the system functions based on the property that some micelle solutions separate phases into a micelle-rich and a micelle-poor phase, depending on conditions such as the temperature, pH, and ionic strength [
31].
6.3. Reverse Aqueous Two-Phase Micellar Systems
“The reverse systems (ATPS-RM) are surfactant-based, using nanometer-sized water pools. These water pools are formed by a monolayer of surfactant molecules entrapping water“ [
32]. These aggregates can solubilise the different molecules depending on their hydrophobicity, size, and charge [
33].
6.4. Ionic Liquid-Based Aqueous Two-Phase Systems
Ionic liquids are also considered green solvents. Ionic liquids have been applied in bio-catalysis, electrochemistry, and bio-separations [
34]. “Ionic liquids have been investigated as novel aqueous two-phase systems (ATPS-IL)” [
35]. Ionic liquids are usually comprised of inorganic or organic cations and anions. Most of the time, they are liquids at room temperature and have low vapour pressure and a wide range of structures.
7. Chromatography Techniques
Chromatography consists of a cluster of separation systems that involve molecular retardation regarding the solvent front that develops over the measurable portion [
36]. Chromatography techniques in separating protein mixtures are the most effective and have been widely used to purify individual proteins [
37]. The purifying process depends on the protein size, charge, hydrophobicity, and bio-specific interaction.
Figure 8 shows the particular properties of the protein [
38].
The first stage of the chromatography procedure is typically a capturing phase, where the product will bind to the adsorbent while the impurities do not [
39]. Further, weakly bound proteins will be washed away so the target protein can be eluted [
40]. There are several types of chromatography methods. The types of chromatography methods are reviewed in the following sections.
7.1. Ion-Exchange Chromatography
Ionic interactions are the basis for protein purification in the ion-exchange chromatography method [
41]. Proteins with different surface charges compete with each other for clusters with opposite directions on an ion-exchanger adsorbent. This increases the separation.
7.2. Affinity Chromatography
Affinity chromatography works based on the principle of an interacting protein. A protein has binding positions with harmonising surfaces to its ligand. “The binding is a mix of van der Waals forces, electrostatic or hydrophobic interactions, and hydrogen bonds” [
42].
8. Conclusions
With further advancements in biotechnology and in-depth research on the structure and function of various proteins, protein separation, and purification technologies, the MCPA system is an amazingly economical way for protein treatment research to purify samples. The new method for purifying proteins helps researchers improve their column chromatography. For example, it helps them figure out what kind and how much resin to use to purify specific proteins and whether to purify proteins on a large scale.
The cells harbouring surface-immobilised SUMO and Ulp1 can be removed by modest centrifugation, with the purified protein confined to the supernatant. Because of the affinity of excess Ulp1 protease for SUMO fusion, excess protein production may be conducted within half an hour using this method. Overall, the cell surface, using the SUMO–Ulp1 approach, provides an easy process for protein purification in its localised shape that requires the optimal cleavage and centrifugation steps.
Regular liquid column chromatography is the most commonly used approach for separating and purifying target proteins and peptides. Magnetic separation strategies are very new and, therefore, under rigorous improvement. Components of magnetic affinity are presently usually used in molecular biology (especially for nucleic acid separation), cell biology, and microbiology (separation of goal cells), and as elements of the techniques for the identification of absolute analyses, the use of magnetic Enzyme Linked ImmunoSorbent Assay (ELISA) and associated strategies (exclusively for the identification of medical markers and environmental impurities). Until now, separations on a small scale have succeeded, and consequently, the full capability of these strategies is no longer suppressed. Overall, magnetic separation strategies can offer a better method of protein purification in the future because of a few advantages.
Use of the ATPS and chromatographic techniques for the downstream processing of recombinant bromelain was investigated. The evaluations have established that the ATPS technique is a cost-effective, time-saving (30 min), and high-recuperation approach that can be scaled up for commercial purposes. Therefore, the ATPS may be a viable single-step separation purification method, moving away from multi-step purification, such as the chromatography technique.
Therefore, it should be pointed out that the purity of the proteins obtained in this review is still insufficient. Consequently, further study and dedication are needed to develop different protein purification methods since intracellular proteins are transported mainly by bacterial cells through the cell membrane. In carrier proteins [
43], the genetic deletion of these proteins will decrease the secretion of non-target proteins; chemicals that prevent protein emission can also obtain protein purity. Even though more research needs to be done before this protein purification method is widely used, this work provides an excellent method to purify proteins.
Author Contributions
Conceptualisation, A.F.P. and M.H.M.S.; methodology, A.F.P. and M.S.N.; validation, M.H.M.S.; investigation, M.H.M.S.; resources, K.J.S., A.F.P. and M.S.N.; data curation, K.J.S., A.F.P. and M.S.N.; writing—original draft preparation, M.H.M.S., K.J.S., A.F.P. and M.S.N.; writing—review and editing, M.H.M.S.; visualisation, A.F.P. and M.S.N.; project administration, M.H.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We highly appreciate our lecturers and friends at the Class of Techniques in Purification and Characterization of Proteins, BCM5704. In addition, we would also like to express our gratitude to our advisors for their assistance and support. Last but not least, we thank the anonymous reviewers for their insights and suggestions to improve this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kimple, M.E.; Brill, A.L.; Pasker, R.L. Overview of Affinity Tags for Protein Purification. Curr. Protoc. Protein Sci. 2013, 73, 9.9.1–9.9.23. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.W. New trends and afnity tag designs for recombinant protein purifcation. Curr. Opin. Struct. Biol. 2014, 26, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, M.; Musante, L.; Ravida, A.; Shortt, B.; Byrne, B.; Holthofer, H. Preparative purification of recombinant proteins: Current status and future trends. BioMed Res. Int. 2013, 2013, 312709. [Google Scholar] [CrossRef] [PubMed]
- Block, H.; Maertens, B.; Spriestersbach, A.; Brinker, N.; Kubicek, J.; Fabis, R.; Labahn, J.; Schäfer, F. Chapter 27 Immobilised-Metal Affinity Chromatography (IMAC). In Guide to Protein Purification, 2nd ed.; Academic Press: Cambridge, MA, USA, 2009; pp. 439–473. [Google Scholar] [CrossRef]
- Stromberg, P.; Rotticci-Mulder, J.; Bjornestedt, R.; Schmidt, S.R. Preparative parallel protein purification (P4). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 818, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C. The process of structure-based drug design. Chem. Biol. 2003, 10, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Safarik, I.; Safarikova, M. Magnetic techniques for the isolation and purification of proteins and peptides. BioMagn. Res. Technol. 2004, 2, 1–17. [Google Scholar] [CrossRef]
- Liu, S.; Li, Z.; Yu, B.; Wang, S.; Shen, Y.; Cong, H. Recent advances on protein separation and purification methods. Adv. Colloid Interface Sci. 2020, 284, 102254. [Google Scholar] [CrossRef]
- Dominguez, M.J.; Lantz, B.J.; Rhode, R.J.; Sharp, Z.L.; Finney, K.C.; Martinez, V.J.; Stollar, E.J. A multi-column plate adapter provides an economical and versatile high-throughput protein purification system. Protein Expr. Purif. 2018, 152, 84–91. [Google Scholar] [CrossRef]
- Zhou, X.F.; Zhang, C.L.; Gao, X.P.; Wang, W.L.; He, Z.F.; Jiang, F.Y.; Pang, Y.L.; Li, J.H.; Ren, X.J.; Zhou, H.B.; et al. Simple and rapid protein purification method based on cell-surface display of SUMO-fused recombinant protein and Ulp1 protease. AMB Express 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Arshad, Z.I.M.; Amid, A.; Yusof, F.; Sulaiman, S.Z.; Mudalip, S.K.A.; Man, R.C.; Shaarani, S.M. Comparison of Purification Methods to Purify Recombinant Bromelain from Escherichia Coli BL21-A1. Malays. J. Anal. Sci. 2017, 21, 958–971. [Google Scholar] [CrossRef]
- Li, B.; Guo, F.; Hu, H.; Liu, P.; Tan, M.; Pan, J.; Zhai, L. The characterisation of column heating effect in nanoflow liquid chromatography mass spectrometry (nanoLC-MS)–based proteomics. J. Mass Spectrom. 2020, 55, e4441. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chen, T.; Iqbal, M.Z.; Yang, F.; Hampp, N.; Wu, A.; Luo, L. Applications of magnetic materials separation in biological nanomedicine. Electrophoresis 2019, 40, 2011–2028. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Kwon, I. Purification-Free, Target-Selective Immobilization of a Protein from Cell Lysates. Biotechnol. J. 2018, 13, 1700739. [Google Scholar] [CrossRef] [PubMed]
- Matte, A. Chapter 9—High-throughput, parallelised and automated protein purification for therapeutic antibody development. In Approaches to the Purification, Analysis and Characterization of Antibody-Based Therapeutics; Matte, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 181–198. [Google Scholar] [CrossRef]
- Kineavy, F.B.; Davies, A.A.; Mitchell, M.R.; Lay, D.; Dominguez, M.J.; Stollar, E.J. An Economical and Versatile High-Throughput Protein Purification System Using a Multi-Column Plate Adapter. JoVE 2021, 171, e62075. [Google Scholar] [CrossRef]
- Lee, S.Y.; Choi, J.H.; Xu, Z. Microbial cell-surface display. Trends Biotechnol. 2003, 21, 45–52. [Google Scholar] [CrossRef]
- Johnson, P.R.; Hochstrasser, M. SUMO-1: Ubiquitin gains weight. Trends Cell Biol. 1997, 7, 408–413. [Google Scholar] [CrossRef]
- Bird, L.E. High throughput construction and small scale expression screening of multi-tag vectors in Escherichia coli. Methods 2011, 55, 29–37. [Google Scholar] [CrossRef]
- Li, J.; Han, Q.; Zhang, T.; Du, J.; Sun, Q.; Pang, Y. Expression of soluble native protein in Escherichia coli using a cold-shock SUMO tag-fused expression vector. Biotechnol. Rep. 2018, 19, e00261. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, L.; Wang, F.; Zhang, J.; Liu, G.; Gao, B.; Wei, D. Novel application of magnetic protein: Convenient one-step purification and immobilisation of proteins. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Schuster, M.; Wasserbauer, E.; Ortner, C.; Graumann, K.; Jungbauer, A.; Hammerschmid, F.; Werner, G. Short cut of protein purification by integration of cell-disrupture and affinity extraction. Bioseparation 2000, 9, 59–67. [Google Scholar] [CrossRef]
- Hofmann, I.; Schnolzer, M.; Kaufmann, I.; Franke, W.W. Symplekin, a constitutive protein of karyo-and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 2002, 13, 1665–1676. [Google Scholar] [CrossRef] [PubMed]
- Asenjo, J.A.; Andrews, B.A. Aqueous two-phase systems for protein separation: A perspective. J. Chromatogr. A 2011, 1218, 8826–8835. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Beg, Q.K.; Khan, S.; Chauhan, B. An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl. Microbiol. Biotechnol. 2007, 60, 381–395. [Google Scholar] [CrossRef]
- Rosa, P.A.J.; Azevedo, A.M.; Aires-Barros, M.R. Application of central composite design to the optimisation of aqueous two—Phase extraction of human antibodies. J. Chromatogr. A 2007, 1141, 50–60. [Google Scholar] [CrossRef]
- Lopes, A.M. Biomolecules extracted by atps: Practical examples. Rev. Mex. De Ing. Química 2014, 13, 359–377. [Google Scholar]
- Taira, A.; Desobgo, Z.S.C.; Nso, J.E. Aqueous Two -Phase System Partial Purification and Characterization of α-Amylase from Burnatia enneandra Micheli. J. Food Stab. 2019, 2, 26–41. [Google Scholar] [CrossRef]
- Ventura, S.P.M.; Santos-Ebinuma, V.C.; Pereira, J.F.B.; Teixeira, M.F.S.; Pessoa, A.; Coutinho, J.A.P. Isolation of natural red colorant from fermented broth using ionic liquid-based aqueous two-phase systems. J. Ind. Microbiol. Biotechnol. 2013, 40, 507–516. [Google Scholar] [CrossRef]
- Rangel-Yagui, C.O.; Pessoa, A., Jr.; Tavares, L.C. Micellar solubilisation of drugs. J. Pharm. Pharm. Sci. 2005, 8, 147–163. [Google Scholar]
- Liu, C.; Nikas, Y.J.; Blankschtein, D. Novel bioseparations using two-phase aqueous micellar systems. Biotechnol. Bioengin. 1996, 52, 185–192. [Google Scholar] [CrossRef]
- Li, S.; Cao, X. Extraction of tea polysaccharides (TPS) using anionic reverse micellar system. Sep. Purif. Technol. 2014, 122, 306–314. [Google Scholar] [CrossRef]
- Storm, S.; Jakobtorweihen, S.; Smirnova, I.V. Solubilisation in mixed micelles studied by molecular dynamics simulations and COSMOmic. J. Phys. Chem. B 2014, 118, 3593–3604. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Han, J.; Wang, Y.; Yan, Y.; Hu, S.; Li, Y.; Zhao, X. Liquid-liquid equilibrium composed of imidazolium tetrafluoroborate ionic liquids sodium carbonate aqueous two-phase systems and correlation at (288.15, 298.15, and 308.15)K. Thermochim. Acta 2011, 523, 221–226. [Google Scholar] [CrossRef]
- Li, Y.; Yang, L.; Zhao, X.; Guan, W. Liquid-liquid equilibria of ionic liquid N-ethylpyridinium tetrafluoroborate+trisodium citrate/ammonium citrate tribasic/sodium succinate/sodium tartrate aqueous two-phase systems at 298.15K. Thermochim. Acta 2012, 550, 5–12. [Google Scholar] [CrossRef]
- Chigome, S.; Torto, N. A review of opportunities for electrospun nanofibers in analytical chemistry. Anal. Chim. Acta 2011, 706, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Jiang, X.; Feng, S.; Tian, R.; Zou, H. Advances in chromatographic techniques and methods in shotgun proteome analysis. TrAC Trends Anal. Chem. 2007, 26, 80–84. [Google Scholar] [CrossRef]
- Hedhammar, M. Strategies for Facilitated Protein Recovery after Recombinant Production in Escherichia coli; School of Biotechnology, Royal Institute of Technology: Stockholm, Sweden, 2005. [Google Scholar]
- Baur, D.; Angarita, M.; Müller-Späth, T.; Steinebach, F.; Morbidelli, M. Comparison of batch and continuous multi-column protein A capture processes by optimal design. Biotechnol. J. 2016, 11, 920–931. [Google Scholar] [CrossRef]
- Hedhammar, M.; Karlström, A.E.; Hober, S. Chromatographic methods for protein purification. Stockh. R. Inst. Technol. 2006, 324, 1–31. [Google Scholar]
- Karlsson, E.; Ryden, L. Ion-Exchange Chromatography. In Protein Purification: Principles, High-Resolution Methods and Applications; Janson, J.C., Ryden, L., Eds.; Wiley: Hoboken, NJ, USA, 1998. [Google Scholar]
- Lee, W.C.; Lee, K.H. Applications of affinity chromatography in proteomics. Analog. Biochem. 2004, 324, 1–10. [Google Scholar] [CrossRef]
- Thanassi, D.G.; Hultgren, S.J. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 2000, 12, 420–430. [Google Scholar] [CrossRef]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).