Abstract
Pathogens-in-Foods (PIF) is a dynamic database constructed using systematic literature searches of occurrence data (prevalence and enumeration) of important pathogenic agents (Bacillus cereus, Campylobacter spp., Clostridium perfringens, Listeria monocytogenes, Salmonella spp., Shiga toxin-producing Escherichia coli, Staphylococcus aureus, Yersinia enterocolitica, Cryptosporidium spp., Giardia spp., Toxoplasma gondii, Hepatitis A virus, Hepatitis E virus and Norovirus) in foods randomly surveyed across Europe. After filtering the primary studies, these were screened for relevance and methodological quality, and the data were extracted into the PIF database following a systematic categorisation of microbiological methods, food types and outcomes. The database is freely accessible through a web application that facilitates data retrieval according to several relevant variables. The PIF spans data published from 2000 onwards and is intended for use by researchers and food authorities after meta-analysis, in microbiological risk assessment.
1. Introduction
In the literature, there are many investigations addressing the identification and quantification of biological hazards in foods surveyed at various stages in the farm-to-fork chain. Being able to access and gather this information has become increasingly relevant in the development of pathogens’ risk assessment models, risk management tools and meta-analysis by both food researchers and food safety authorities. Nevertheless, this information is largely dispersed, disharmonised and not always accessible. To this end, the Pathogens-in-Foods (PIF) database was created to bring together, under a harmonised arrangement, methodologically sound data on the prevalence and enumeration of relevant pathogens occurring in different food matrices produced, commercialised, and/or consumed in Europe. The PIF was constructed to facilitate the access, visualisation, and assessment of microbiological occurrence data from different sources. The objective of this paper was to demonstrate the basis and utility of the database and highlight the resourcefulness of its web application interface.
2. Systematic Review and Extraction to PIF
According to EFSA’s guidelines [1], a systematic review is “an overview of existing evidence pertinent to a clearly formulated question, which uses pre-specified and standardised methods to identify and critically appraise relevant research, and to collect, report and analyse data from the studies that are included in the review”.
Based upon these guidelines, the research group developed a systematic review protocol to be implemented prior to the review, starting with the definition of a focused review question that helped define the most relevant terms for literature search. The resulting list of candidate studies were screened for relevance to the review question, and subsequently, the methodological quality of the studies was assessed using the pre-set quality criteria. After validation, the qualitative and quantitative data pre-determined in the protocol was extracted and fed onto the database.
The database contains data extracted from the year 2000 onwards [2], but for the purpose of this paper, the authors highlight the last systematic review applied to papers published between 2020 and 2022.
2.1. Definition of the Review Question
The definition of the review question followed the PO question structure defined by EFSA [1] that is typically applied to questions about occurrence in a given population (P) of a certain outcome (O). In this case, the population was determined as foods commercialised in Europe, while the outcome encompassed the most relevant foodborne pathogens.
2.2. Literature Search
In December 2021, systematic literature searches were conducted on bibliographic engines PubMed®, SciELO, Scopus®, and Web of Science™, using a selection of included and excluded keywords to create a search query adapted to each engine. Key terms included the most important biological hazards (Bacillus cereus, Campylobacter spp., Clostridium perfringens, Listeria monocytogenes, Salmonella spp., Shiga toxin-producing Escherichia coli, (STEC; VTEC, EHEC; O157, O157:H7, O26:H11, O145:H28, O103:H2, O111:H8, O104:H4) Staphylococcus aureus, Yersinia enterocolitica, Cryptosporidium spp., Giardia spp., Toxoplasma spp., Hepatitis A virus, Hepatitis E virus, and Norovirus), a list of several food matrices (among them meat and meat products, egg and egg products, milk and dairy, seafood and fishery products, produce, fruits, ready-to-eat, composite and multi-ingredient foods, oils, sugars, grains, and beverages), additional terms (including, but not limited to occurrence, prevalence, incidence, presence, contamination, survey, sampling, and “microbiological quality”), and excluded terms associated with artificial contamination, challenge studies or meta-analysis, among others. Search queries were constructed using the defined key terms interspersed with the appropriate Boolean operators AND, OR and NOT, adapted to each bibliographic engines’ language and set to search for these terms in title/abstract/keywords only. Whenever the engine filters allowed, searches were limited to peer-reviewed articles and reports which took place in European countries, published in English, Spanish, French, and/or Portuguese, between 2020 and 2022.
The references of the filtered studies were extracted in BibTeX file format from each search engine and after combining the files using the JabRef v. 5.6 reference manager software [3], the joined raw file was cleaned of duplicates. Table 1 presents the number of references extracted by bibliographic search engine and the total number of citations after duplicate removal. In most cases, studies were duplicated or even triplicated across the main three resulting engine searches, which accounts for the lower record post duplicate cleaning.

Table 1.
Number of records retrieved by search engine and the total number of records with and without duplicates.
To consolidate data regarding “drinking water”, a second systematic literature search was performed in January 2022 following the same procedure but using “drinking water”-related search terms in the search queries. For this search, Scopus and PubMed were the only bibliographic engines used. In the previous search, results from Web of Science were mostly duplicates of studies retrieved with the other two engines, and as for SciELO, this engine retrieved mostly studies carried out in Latin America, which were out of scope.
2.3. Screening for Relevance and Methodological Quality Assessment
The cleaned BibTeX file was uploaded to the Rayyan systematic review web tool [4], where each study was screened for relevance individually by two researchers. The title and abstract of every reference were assessed based on its ability to answer the review question according to the following criteria: (i) investigation of foods either produced or commercialised/consumed in European countries, (ii) occurrence of any of the target foodborne hazards in said foods, (iii) non-outbreak related occurrence, (iv) randomised sampling, (v) investigation of naturally contaminated food matrices (no experimental contamination), and (vi) investigation of non-treated controls in experimental studies. Entries were either marked as “included” if they met the established criteria and “excluded” if not. Whenever the assessment was inconclusive based on title/abstract reviewing and the study was marked “maybe”, or when there was a conflict between the two researchers’ decisions, the final ruling for inclusion/exclusion was determined by a third researcher. Validated studies were extracted from Rayyan and re-uploaded to JabRef, where a “Citationkey” identifier (StudyID) was attributed to each reference (FirstAuthorSurname_JournalAbbreviation_YearofPublication), and full texts were appended to each respective entry.
Afterwards, two researchers read the full texts of the primary studies, in order to further appraise their suitability for inclusion in the database, and then carried out the methodological quality assessment, following a standardised checklist of criteria: (a) detection/quantification of biological hazards by approved/well described microbiological methods (sample weight, microbiological media or techniques used); (b) sufficient data on prevalence (sample size and number of contaminated samples) and enumeration (sample size, limit of quantification, mean and standard deviation); and (c) clear food classification and food chain information of the studied samples. The studies that did not meet one of the criteria were not necessarily rejected, but instead were marked as “potentially biased”. If more than one criterion was not met, the study was discarded. The final decision of removing a primary study was determined by the rejection of the third researcher. When relevant, the bibliographic references of primary studies were screened for additional eligible articles, and “new” references were manually added to the database.
2.4. Data Extraction
After validating the methodological quality, the data were extracted into separate sections built for each of three major pathogen groups (bacteria, viruses, or parasites) in the PIF database, following a pre-determined built-in systematic categorisation.
In the first section, primary study characteristics like StudyID, type of study (survey/comparison/other), country of publication, duration, and year of study were added. Next, pathogen information was uploaded, specifically pathogen identification and other specific bacteria (serotype/serovar), virus (genotype/sub-genotype) or parasite (species/subtype) data when applicable, followed by microbiological methods, namely if the study reports prevalence/count or both type data, nature of the assay (culture, DNA, immuno-based, microscopy or others) and respective method/technique used. For studies with viruses or parasites, further details may have been added if the study contemplated an identification or infectivity assay, or even specific sample preparation for parasites (i.e., centrifugation, flotation, etc.).
A second section, required all food and food chain characteristics, including food category (beverages, composite, dairy, eggs, fruits, grains, legumes, meat, oils, seafood, sugars, and vegetables), a sub-hierarchy for every food category, species (in the case of animal origin foods), food processing class (minced, pre-cut, pasteurised, cooked, cured, marinated, smoked, raw, minimally processed, dried, fermented, or not applicable-NA) and other subcategory-specific information. An additional requirement is the packaging status of food (packed, unpacked, various, NA), stage in the food chain (farm, mid-processing, end-processing, retail, restauration), temperature class at retail (ambient, chilled, frozen, various, NA) and ready-to-eat (RTE) status (yes/no). For viruses and parasites, given the specific nature of certain detection methods used, the sampled organ may be detailed further in studies focusing on seafood molluscs (digestive tissues, mantle, gills, whole flesh, or not-specified-N/S) or meat (diaphragm, liver, heart, brain, blood, or meat juice).
The third and last section of the database pertains to all prevalence and/or enumeration results. For bacteria, required fields included sample weight and unit, sample size, confirmation of pathogen status (yes/no) and potential-for bias status (yes/no), as well as prevalence data regarding the number of enriched samples, or limit of quantification (LoQ) and number of samples above and below LoQ, in cases of enumeration. Other data could be uploaded such as limit of detection (LoD), histogram of frequencies for counts, maximum counts, mean microbial concentration and standard deviation.
For parasites and virus, counts units (raw, Log10 or Ln) and mean concentration were required fields for enumeration. Specific data regarding the quality of the detection method (nature of the control virus, extraction efficiency, etc.) could be added for viruses and infectivity results for both groups.
3. Overview
Presently, the Pathogens-in-Foods database includes 1153 primary studies, with over 5200 bacteria, 200 virus, and 40 parasite entries, spanning data published from 2000 onwards to the present day. Systematic reviews were conducted periodically through the same process, to ensure that new data was continuously added, and the database is kept up to date.
The PIF is easily accessible through the main page at https://fsqa.esa.ipb.pt/, and the following case study provides an example on how to retrieve the data and its applicability. To retrieve data on the occurrence (prevalence and counts) of L. monocytogenes in non-RTE frozen vegetables sampled at the end of processing and at retail, PIF must first be accessed through the “Access System” function on the right side of the main page (Figure 1a).
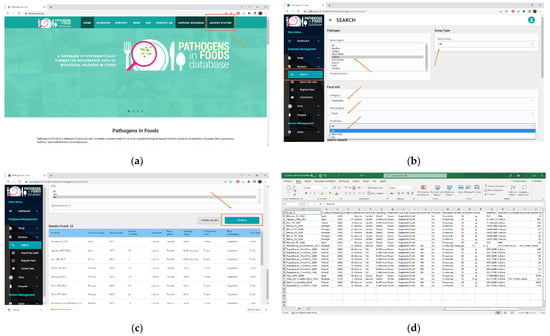
Figure 1.
(a,b) Accessing the PIF database and (c,d) retrieval of data.
After login, using the “Search” function on the “Bacteria” dropdown menu on the left side of the screen opens the page where variables of the search can be defined. In the first section “Listeria” should be chosen as the pathogen of interest, and the type of assay should be marked as “all”. Next, food characteristics can be filtered according to the category (“vegetables”), sub-category (“fresh”) and food class (“all”) (Figure 1b). Through advanced filters, further features such as country of food origin (“all”), StudyID, label, packaging status (“all”), sampling stage (“endprocessing” and “retail”), temperature at retail (“frozen”) and RTE status (“no”) can be included in the search. Search results can be presented as a summarised table in the database interface or downloaded as CVS or JSON format files (Figure 1c,d). The extracted data file contains all the previously detailed information that has been extracted from the primary studies, each line corresponding to a different food sample. The available data can, for example, be constructed into a table reporting prevalence across multiple countries, like the one detailed by Table 2.

Table 2.
Listeria. monocytogenes prevalence and counts (when available) in non-RTE frozen vegetables sampled at the end of processing and at retail.
4. Conclusions
The Pathogens-in-Foods database and the associated web application are highly intuitive and easy to use, providing in depth detection and enumeration data, while also generating dynamic graphs and summary statistics of incidence through interactive dashboards. The PIF is intended to be open with free access for researchers and risk assessment organisations, providing them with a tool that compiles reliable, and quality assessed data for quantitative microbiological risk assessment.
Author Contributions
Conceptualization, U.G.-B., M.S., P.K. and V.C.; methodology, A.S.F., M.W., U.G.-B. and V.C.; software, M.S., P.K., A.T., U.G.-B. and V.C.; validation, U.G.-B., L.G. and V.C.; formal analysis, A.S.F., M.W., V.C. and U.G.-B.; investigation, A.S.F. and M.W.; resources, M.S., P.K., A.T., U.G.-B. and V.C.; data curation, A.S.F., M.W. and U.G.-B.; writing—original draft preparation, A.S.F. and U.G.-B.; writing—review and editing, A.S.F. and U.G.-B.; visualization, L.G., M.S., P.K., A.T., A.S.F., U.G.-B. and V.C.; supervision, U.G.-B. and V.C.; project administration, U.G.-B., M.S., P.K. and V.C.; funding acquisition, M.S., P.K., U.G.-B. and V.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the French Agency for Food, Environmental and Occupational Health and Safety (ANSES), Grant Agreement ANSES/IPB Nº2021-CRD-03 and by the European Food Safety Authority (EFSA), Grant Agreement GP/EFSA/BIOHAW/2022/01 (since 1 September 2022). The authors are grateful to the Foundation for Science and Technology (FCT, Portugal) for financial support through national funds FCT/MCTES (PIDDAC) to CIMO (UIDB/00690/2020 and UIDP/00690/2020) and SusTEC (LA/P/0007/2021). Gonzales-Barron acknowledges the national funding by FCT, P.I., through the Institutional Scientific Employment Programme contract.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available from the Pathogens-in-Foods database: https://fsqa.esa.ipb.pt/overview/ (accessed on 15 September 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- European Food Safety Authority (EFSA). Application of systematic review methodology to food and feed safety assessments to support decision making. EFSA J. 2010, 8, 1637. [Google Scholar] [CrossRef]
- Deusdado, S.; Cadavez, V.; Rodrigues, V.; Kooh, P.; Sanaa, M.; Gonzales-Barron, U. Pathogens-in-foods: A database of occurrence of microbiological hazards in foods commercialised in Europe. In Proceedings of the AGROSTAT 2018, Aix-Marseille University-ISM2, Marseille, France, 14–16 March 2018. [Google Scholar]
- JabRef Development Team. JabRef—An Open-Source, Cross-Platform Citation and Reference Management Software. Version 5.1. 2021. Available online: https://www.jabref.org (accessed on 15 September 2022).
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Aguado, V.; Vitas, A.I.; Garcia-Jalon, I. Characterization of Listeria monocytogenes and Listeria innocua from a vegetable processing plant by RAPD and REA. Int. J. Food Microbiol. 2004, 90, 341–347. [Google Scholar] [CrossRef]
- Pappelbaum, K.; Grif, K.; Heller, I.; Wüirzner, R.; Hein, I.; Ellerbroek, L.; Wagner, M. Monitoring hygiene on- and at-line is critical for controlling Listeria monocytogenes during produce processing. J. Food Prot. 2008, 71, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Cetinkaya, F.; Soyutemiz, G.E. Occurrence of Listeria species in the processing stages of frozen pepper. J. Food Saf. 2007, 27, 134–147. [Google Scholar] [CrossRef]
- Skowron, K.; Grudlewska, K.; Lewandowski, D.; Gajewski, P.; Reśliński, A.; Gospodarek-Komkowska, E. Antibiotic susceptibility and ability to form biofilm of Listeria monocytogenes strains isolated from frozen vegetables. Acta Aliment. 2019, 48, 65–75. [Google Scholar] [CrossRef]
- Vitas, A.I.; Garcia-Jalon, V.A. Occurrence of Listeria monocytogenes in fresh and processed foods in Navarra (Spain). Int. J. Food Microbiol. 2004, 90, 349–356. [Google Scholar] [CrossRef]
- Mena, C.; Almeida, G.; Carneiro, L.; Teixeira, P.; Hogg, T.; Gibbs, P.A. Incidence of Listeria monocytogenes in different food products commercialized in Portugal. Food Microbiol. 2004, 21, 213–216. [Google Scholar] [CrossRef]
- Vojkovská, H.; Myšková, P.; Gelbíčová, T.; Skočková, A.; Koláčková, I.; Karpíšková, R. Occurrence and characterization of food-borne pathogens isolated from fruit, vegetables and sprouts retailed in the Czech Republic. Food Microbiol. 2017, 63, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Moravkova, M.; Verbikova, V.; Michna, V.; Babak, V.; Cahlikova, H.; Karpiskova, R.; Kralik, P. Detection and Quantification of Listeria monocytogenes in Ready-to-eat Vegetables, Frozen Vegetables and Sprouts Examined by Culture Methods and Real-time PCR. J. Food Nutr. Res. 2017, 5, 832–837. [Google Scholar] [CrossRef]
- Willis, C.; McLauchlin, J.; Aird, H.; Amar, C.; Barker, C.; Dallman, T.; Elviss, N.; Lai, S.; Sadler-Reeves, L. Occurrence of Listeria and Escherichia coli in frozen fruit and vegetables collected from retail and catering premises in England 2018–2019. Int. J. Food Microbiol. 2020, 334, 108849. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).