Abstract
We address the application of phytopathogen filtrates to induce an immune response on plants that may protect them from disease. We exposed Arabidopsis thaliana plants to filtrates of necrotrophic and biotrophic phytopathogens and evaluated whether these triggered an immune response correspondent to each pathogen’s infection pathway. We show that filtrates induce a systemic immune response on plants, but this was not specific to the infection type of phytopathogens. When facing a real infection, however, the filtrates enhanced the immune response compared to the control plants. Moreover, the filtrates increased plant growth by acting either as fertilizers or chemical inducers. Our study demonstrates the biotechnological potential of phytopathogen filtrates.
1. Introduction
Phytopathogen infection on crops decreases the yield and quality of agricultural production, generating considerable economic losses and reducing food security worldwide [1,2,3]. Considerable efforts have been made to counteract phytopathogens with chemical compounds (i.e., bactericides and fungicides). However, besides often being deleterious to ecosystems, chemicals have the disadvantage of inducing pathogen resistance over time [4]. A valuable alternative may be making plants less susceptible to pathogens by an induced immune resistance [5,6]. Induced resistance consists of sensitizing the plant to activate its defense mechanisms by an elicitor agent and preparing the plant for the pathogen arrival, infection, and colonization [7,8]. The pathogen filtrates may be able to activate the defense system in plants because they contain specific molecules, such as proteins, oligosaccharides, oligopeptides, and toxins,, which are detected by receptors in the plant cuticle and trigger a microorganism recognition signature [4,8,9,10,11,12].
The induced resistance response to filtrates should be specific to the microorganism’s biology [13] and their interaction with the host [14]. For instance, biotrophic pathogens suppress the host’s immune system and derive nutrients from living cells, whereas necrotrophic pathogens secrete toxins in order to rapidly kill the host’s tissues and thrive on dead tissues. Hemibiotrophic pathogens combine both strategies of nutrient acquisition, starting with a biotrophic phase followed by a necrotrophic phase [15]. Plants can fight back biotrophic or necrotrophic pathogens through the balanced interaction between the phytohormones of the signaling pathways that mainly include salicylic acid (SA) against biotrophic pathogens and jasmonic acid (JA) against necrotrophic pathogens [13,16].
Here, we assessed the immune response of Arabidopsis thaliana to filtrates of biotrophic, hemibiotrophic, and necrotrophic phytopathogens. These filtrates elicit the expression of a specific immune pathway. We thus assessed whether the biotrophic, hemibiotrophic, and necrotrophic pathogens prompted the expression of genes associated with the JA and SA pathways, respectively. The induced resistance should enhance the defensive response of plants when facing a real infection. Thus, we infected plants with the necrotic fungus Botrytis cinerea and expected the expression of defense genes to be highest in the plants that were exposed to the filtrates from necrotrophic microorganisms. Finally, we assessed whether inducing a sustained immune response with filtrates had the trade-off of reducing plant growth and production.
2. Materials and Methods
We used wild-type A. thaliana Columbia background (Col-0) plants obtained from the Arabidopsis Information Service (AIS) (https://www.arabidopsis.org, accessed on 31 October 2019). Seeds of A. thaliana were surface sterilized and plated on Murashige and Skoog (MS) medium (Sigma-Aldrich, Darmstadt, Germany) [17], solidified with 1% agar (Chem-Lab, Zedelgem, BE) (w/v), and supplemented with 1% sucrose (Sigma-Aldrich, Darmstadt, Germany) (w/v). Seeds were incubated for 7 days at 22 °C with a long-day photoperiod (16 h of light) and 40–50% relative humidity (RH). Seedlings were then transferred to a solid substrate of peat and vermiculite (3:1) and were kept in the greenhouse at 22 °C and 60% RH.
We used bacteria, fungi, and an oomycete as sources of microorganism filtrates. Pectobaterium carotovorum, Rhizoctonia solani, Sclerotinia sclerotiorum, and Phytium irregulare were the necrotrophic phytopathogens. Whereas the species Pseudomonas syringae pv. Tomato and Fusarium oxysporum f.sp. conglutinans were the biotrophic and hemibiotrophic phytopathogens, respectively. Microorganisms were wild strains isolated from infected crops by the Centro Regional de Diagnóstico de Aldearrubia (Junta de Castilla y León, Salamanca, Spain). The bacteria (P. syringae and P. carotovorum) were grown on solid Luria-Bertani (LB) medium (Sigma-Aldrich, Darmstadt, Germany) [18] at 28 °C, while fungi (F. oxysporum f. sp. conglutinans, S. sclerotiorum, and R. solani) and oomycete (P. irregulare) were grown on potato dextrose agar (PDA) medium (Oxoid, Hampshire, UK) at 25 °C. After 7 days, the cultures were diluted in 5 mL of sterile distilled water to obtain a suspension with an optical density between 0.15 and 0.19, except for F. oxysporum, with which we used a 2.3 × 103 spores mL−1 suspension. Bacteria suspensions were cultivated in LB liquid medium and fungi and the oomycete were cultivated in potato dextrose broth (PDB) medium (Neogen, Scotland, UK). Both cultures were incubated with orbital shaking at 180 rpm, and at 28 °C for 48 h. Mediums were then filtrated through 0.22 μm Millipore filters, sealed, and stored at −20 °C.
We used the necrotrophic fungus Botrytis cinerea B05.10 strain as an infection agent, provided by the Phytopathology and Biological Control Group of the Instituto Hispano Luso de Investigaciones Agrarias (CIALE), Spain. B. cinerea was grown in PDA medium (Oxoid, Hampshire, UK) at 25 °C for 7 days, after which the culture was diluted in 5 mL of sterile distilled water to obtain a suspension with 2 × 107 spores mL−1.
In Planta Essays
We evaluated the effect of filtrates from six phytopathogens on the defense gene expression before and after an infection with B. cinerea, as well as plant growth and production. We applied 400 µL of each phytopathogen filtrate on the substrate of 30 A. thaliana plants (~2 cm-long), not further than 0.5 cm from the stem (30 plants × 6 filtrate types). We applied distilled water to another 30 plants, which served as controls.
We assessed the induced immune response by estimating the expression of genes associated with the JA and SA signaling pathways. We collected leaves and roots of nine plants of each filtrate treatment ten days after filtrate application and stored each tissue separately at −80 °C in liquid nitrogen. At the same time, six plants in each filtrate treatment were infected with B. cinerea. We applied 5 μL of B. cinerea spore suspension on three leaves of each plant and sealed them inside of a plastic box for 15 days in a growth chamber at 22 °C, 40% RH, and a 16 h light/8 h dark photoperiod at 80–100 μE m−2 s−1. Infected leaves were collected and stored at −80 °C in liquid nitrogen (n = 6 × 7). We pooled stored tissue samples of each filtrate and infection treatment (nine leaf/root samples in non-infected treatments, and 6 samples in infected treatments) to maximize the amount of genetic material analyzed. We extracted total RNA from the leaf and root samples using the Trizol method (Thermo Fisher Scientifics, Waltham, MS, USA) following the commercial protocol. We used PrimeScriptTM RT reagent kit (Takara, Göteborg, SWE) to synthesize complementary DNA from RNA. We used real-time PCR using a StepOnePlus Applied Biosystems equipment with the KAPA SYBR® FAST qPCR Master Mix Kit (2X) ABI Prism (KAPABiosystems, Cape Town, South Africa) and primers to amplify ICS1 and PR1 genes associated with the SA pathway, and LOX1 and VSP2 associated with the JA pathway, and Actin endogenous gene to assess a baseline genetic expression (Table A1). We applied the PCR program as described in Poveda, 2018 [19]. The resulting threshold cycle values (Ct) of gene amplification were analyzed using the delta–delta Ct method to assess the expression of SA and JA pathway genes relative to the expression of the endogenous gene and relative to the control treatment [20]. We calculated the averages of three Ct value calculations, resulting in one unique value of gene expression per gene, tissue (leaves or roots), and filtrate/infection treatment.
We also evaluated the effect of filtrates on plant growth and seed production. Seventy days after filtrate application, we assessed the effects of the filtrates on plant growth on seven plants per treatment (n = 7 plants × 7 treatments = 49 aerial/root growth measurements). To do this, we removed the plants from the substrate and cleaned the roots. We cut separate roots from aerial tissue and measured their dry weight after being placed in an oven at 65 °C for 48 h. Finally, we waited until the eight plants per treatment fructified (100 days after sowing), and we counted the siliques to assess the effects of the filtrates on plant yield (n = 8 plants × 7 treatments = 56 production measurements). We compared the root weight, aerial weight, and silique number between the filtrate treatments using three general linear models.
All statistical analyzes were carried out in R software [21]. The packages ggplot2 [22], ggpubr [23], and lemon [24] were used for the design of the figures.
3. Results
The phytopathogen filtrates induced the expression of defense genes in A. thaliana up to ten times more than in the control plants (Figure 1). However, not all of the phytopathogen filtrates enhanced gene expression to the same degree in roots and leaves. Most of the gene induction in the leaves concentrated on LOX1 (JA pathway) in plants exposed to filtrates of P. carotovorum, F. oxysporum, and P. irregulare (Figure 1). In the roots, most of the gene induction concentrated on ICS1 (SA pathway) by P. irregulare, P. syringae, and F. oxysporum (Figure 1). A. thaliana leaves infected with B. cinerea mostly induced the gene expression of either PR1 (SA pathway) or VSP2 (JA pathway), whereas the expression of ICS1 and LOX1 genes was minimal. However, the plants expressed either PR1 or VSP2, but never both to the same degree (Figure 1).
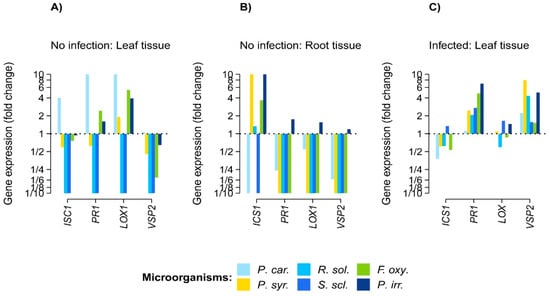
Figure 1.
Effects of phytopathogen filtrates on the expression of Arabidopsis thaliana defense genes before (A,B) and after (C) infection by Botrytis cinerea. Gene expression is relative to endogenous gene expression and relative to control plants using delta–delta Ct units log10 transformed representing proportional change. This shows the necrotrophic phytopathogens in blue, hemibiotrophic in green, and biotrophic in yellow. Microorganisms correspond as follows: P. car.: Pectobacterium carotovorum, P. syr.: Pseudomonas syringae, R. sol.: Rhizoctonia solani, S. scl.: Sclerotinia sclerotiorum, F. oxy.: Fusarium oxysporum, and P. irr.: Pythium irregulare.
The plants that were exposed to the filtrate treatments exhibited greater aerial and radicular biomass compared to the control plants. However, the plants that were exposed to S. sclerotiorum filtrate were not different from the control plants (Table A2, Figure 2). Regarding fruit production, there were no differences between the plants that were treated with the filtrates and the control plants (Table A2, Figure 2).
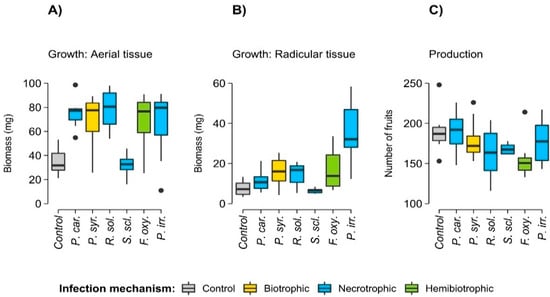
Figure 2.
Effect of phytopathogen filtrates on the growth (A,B) and silique production (C) of Arabidopsis thaliana plants. Microorganisms correspond as follows: P. car.: Pectobacterium carotovorum, P. syr.: Pseudomonas syringae, R. sol.: Rhizoctonia solani, S. scl.: Sclerotinia sclerotiorum, F. oxy.: Fusarium oxysporum, and P. irr.: Pythium irregulare.
4. Discussion
We have shown that the inoculation of A. thaliana rhizosphere with phytopathogen filtrates induced the gene expression of the SA and JA pathways, in radicular and aerial tissue, suggesting the activation of a systemic defensive response [25]. This adds to the body of literature showing that the filtrates contain chemical compounds from the pathogens that the plants recognize despite the absence of the living microorganisms [26,27]. Pathogen-derived elicitors can trigger a plant’s immune response by activating a signaling cascade communicated throughout the plant [25]. Here, local immune responses induce mobile signals that reach towards the distal tissues in order to initiate a secondary immune response [28], conferring an enhanced resistance against subsequent infections, which has been referred to as systemic acquired resistance (SAR) [29]. For instance, we have shown that the filtrates enhanced the immune response against an infection from B. cinerea compared to control plants. Thus, triggering this process through chemical elicitors in microorganism filtrates may be used as a tool to prepare a plant’s immune system against real disease, akin to a vaccination [30].
We have found, however, that filtrates from necrotrophic and biotrophic microorganisms did not induce an exclusive expression of JA and SA pathways respectively, suggesting a plant-induced immune response by microorganism filtrates was not specific. This may result from plants being able to express both SA and JA pathways simultaneously, as a preventive strategy against an infection, that has not been fully identified [31]. Alternatively, but not exclusively, a non-specific immune response may be a consequence of phytopathogens infection strategies, some of which ‘trick’ plants into committing to a defense pathway that is not effective against the pathogen [32]. For instance, P. syringae induced the expression of VSP2 despite being a biotrophic phytopathogen, which may result from its infection mechanism that secretes coronatine, which ‘tricks’ the host-plant into committing to a JA defense response instead of the corresponding SA response [33]. Filtrates likely contain an array of pathogen signals that may prompt a non-specific immune response [31], and it has been suggested that specific defense pathways can only be triggered through the complex interaction between the pathogen and its host [25]. Paradoxically, studying microorganism filtrates is a promising tool to understand the molecular mechanism of plant defensive responses [12,34,35], which may help developing microorganism-derived filtrates that are specifically tailored to fight a target disease.
The exposure to the phytopathogen filtrates enhanced plant growth, on top of inducing an immune response, despite a well-known trade-off between plant growth and the defensive response [36]. Free-living microbes, and a variety of plant growth-promoting rhizobacteria (PGPR), are able to stimulate plant growth by different direct or indirect mechanisms, such as the production of phytohormones, decomposition, mineralization of organic material, and enhancing the bioavailability of mineral nutrients [37,38]. For instance, the volatile components in phytopathogen filtrates have been shown to increase plant growth and fruit production in pepper plants [39]. However, we did not find an increase in production in our study, which may be caused by the specific signaling pathways of each phytopathogen and each plant [40,41].Thus, the mixture of components constituting phytopathogen filtrates may act simultaneously as immune response elicitors, as well as fertilizer [42].
5. Conclusions
Our study demonstrated that phytopathogen filtrates contain chemical signals that can trigger a systemic immune response in plants. This response, however, was not specific to the infection mechanism of the phytopathogen from which the filtrate was extracted. Still, filtrates did bolster the plants’ immune response when facing a real infection. Moreover, we observed that the filtrates stimulated plant growth without causing a trade-off with their immune response. Thus, our study provides evidence that phytopathogen filtrates may be used to enhance the immune response of plants against a real infection. Further research could focus on developing filtrates that are tailored to elicit specific defense pathways. The specific responses that filtrates may trigger in plants (e.g., activation of enzymes and crosstalk of phytohormones pathways, among others) may hold great agrobiotechnological potential.
Supplementary Materials
The video presentation can be downloaded at the following address: https://www.mdpi.com/article/10.3390/IECPS2021-11974/s1.
Author Contributions
A.C.Á. conducted experiments, recorded data, performed analyses, and wrote the manuscript. J.P. designed the experiments and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was part of Ávila A.C. M.Sc. program funded by Banco Santander and the Universidad de Salamanca under the scholarship “Becas internacionales para la movilidad en estudios de máster 2019–2020”. Research work was funded by Grupo de Fitopatología y Control Biológico of the Instituto Hispanoluso de Investigaciones Agrarias (CIALE).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Mendeley Data at the following doi:1017632/y3b226n5v3.1.
Acknowledgments
The authors would like to thank Luis F. Camacho for his invaluable support in the development of this research. We also thank all the people that constitute the Grupo de Fitopatología y Control Biológico (CIALE, Universidad de Salamanca, Salamanca, Spain).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Oligonucleotides used in gene expression analysis in A. thaliana.
Table A1.
Oligonucleotides used in gene expression analysis in A. thaliana.
| Gene | Application | Sequences (5′-3′) |
|---|---|---|
| ICS1 | SA synthesis | GATCTAGCTAACGAGAACGG |
| ICS1 | SA synthesis | CATTAAACTCAACCTGAGGGAC |
| PR1 | SA response | CAAAGTGAGGTGTAACAATGGTGGA |
| PR1 | SA response | ATGGCTTCTCGTTCACATAATTCCC |
| LOX1 | JA synthesis | TCAACGATTTCAATGCTTCGTTTCT |
| LOX1 | JA synthesis | TCAGAGCTTACAAGACGAAGAGTG |
| VSP2 | JA response | GTTAGGGACCGGAGCATCAA |
| VSP2 | JA response | TCAATCCCGAGCTCTATGATGTT |
| Actin | Endogenous gene | CTCCCGCTATGTATGTCGCC |
| Actin | Endogenous gene | TTGGCACAGTGTGAGACACAC |

Table A2.
Statistical estimates of general linear models comparing the root dry-weight, aerial dry-weight, and silique production of A. thaliana plants exposed to different phytopathogen filtrates. Estimate values reflect means for each filtrate treatment compared to the control treatment.
Table A2.
Statistical estimates of general linear models comparing the root dry-weight, aerial dry-weight, and silique production of A. thaliana plants exposed to different phytopathogen filtrates. Estimate values reflect means for each filtrate treatment compared to the control treatment.
| Response Variable | Fixed Factor | Estimate ± S.E. | F | p Value |
|---|---|---|---|---|
| Root dry-weight (log10 transformed mg) (n = 49) | Phytopathogen filtrate: | 5.79 | <0.01 | |
| F. oxysporum | 0.41 ± 0.11 | |||
| P. carotovorum | 0.15 ± 0.11 | |||
| P. irregulare | 0.46 ± 0.11 | |||
| P. syringae | 0.23 ± 0.11 | |||
| R. solani | 0.32 ± 0.11 | |||
| S. sclerotiorum | −0.01 ± 0.11 | |||
| Aerial dry-weight | Phytopathogen filtrate: | 8.91 | <0.01 | |
| (n = 49) | F. oxysporum | 0.03 ± 0.01 | ||
| P. carotovorum | 0.03 ± 0.01 | |||
| P. irregulare | 0.02 ± 0.01 | |||
| P. syringae | 0.02 ± 0.01 | |||
| R. solani | 0.03 ± 0.01 | |||
| S. sclerotiorum | 0.01 ± 0.01 | |||
| Number of siliques | Phytopathogen filtrate: | 2.15 | <0.06 | |
| (n = 56) | F. oxysporum | −34.2 ± 12.7 | ||
| P. carotovorum | 0.5 ± 12.7 | |||
| P. irregulare | −13.2 ± 12.7 | |||
| P. syringae | −10.6 ± 12.7 | |||
| R. solani | −26.6 ± 12.7 | |||
| S. sclerotiorum | −22.1 ± 12.7 |
References
- Binyamin, R.; Nadeem, S.M.; Akhtar, S.; Khan, M.Y.; Anjum, R. Beneficial and pathogenic plant-microbe interactions: A review. Soil Environ. 2019, 38, 127–150. [Google Scholar] [CrossRef]
- Kamoun, S.; Wu, C.H.; Derevnina, L. Receptor networks underpin plant immunity. Science 2018, 360, 1300–1301. [Google Scholar]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, Y.; Li, Y.; Dong, J.; Liu, X.; Li, C. Biocontrol of Rhizoctonia solani via Induction of the Defense Mechanism and Antimicrobial Compounds Produced by Bacillus subtilis SL-44 on Pepper (Capsicum annuum L.). Front. Microbiol. 2019, 10, 02676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, M.; Sohal, B.S. Role of Elicitors in Inducing Resistance in Plants against Pathogen Infection: A Review. ISRN Biochem. 2013, 2013, 762412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuc, J. Induced immunity to plant disease. BioScience 1982, 32, 854–860. [Google Scholar]
- Conrath, U.; Pieterse, C.; Mauch-Mani, B. Priming in plant–pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef] [Green Version]
- da Silva, A.; Freitas, K.; Bastidas, J.; dos Santos, M.; Lilianne, M. Induction of defense mechanisms from filtrates of saprophytic fungi against early blight disease in tomato. Afr. J. Microbiol. Res. 2016, 10, 1849–1859. [Google Scholar]
- Dubery, I.; Sanabria, N.; Huang, J. Nonself perception in plant innate immunity. Adv. Exp. Med. Biol. 2012, 738, 79–107. [Google Scholar]
- Bae, S.J.; Mohanta, T.K.; Chung, J.Y.; Ryu, M.; Park, G.; Shim, S.; Hong, S.B.; Seo, H.; Bae, D.W.; Bae, I.; et al. Trichoderma metabolites as biological control agents against Phytophthora pathogens. Biol. Control 2016, 92, 128–138. [Google Scholar] [CrossRef]
- Stracquadanio, C.; Quiles, J.M.; Meca, G.; Cacciola, S.O. Antifungal activity of bioactive metabolites produced by trichoderma asperellum and trichoderma atroviride in liquid medium. J. Fungi 2020, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; da Silva, A.; Bastidas, J.; Pascholati, S.; Freitas, K. Induction of defense mechanisms in tomato plants by saprobic fungi filtrates against early blight disease1. Rev. Caatinga 2020, 33, 671–678. [Google Scholar]
- Malik, N.A.A.; Kumar, I.S.; Nadarajah, K. Elicitor and receptor molecules: Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, S.; Basu, A.; Kundu, S. Biotrophy-necrotrophy switch in pathogen evoke differential response in resistant and susceptible sesame involving multiple signaling pathways at different phases. Sci. Rep. 2017, 7, 17251. [Google Scholar] [CrossRef] [Green Version]
- Panthapulakkal, S.; Lung, S.C.; Liao, P.; Lo, C.; Chye, M.L. The overexpression of OsACBP5 protects transgenic rice against necrotrophic, hemibiotrophic and biotrophic pathogens. Sci. Rep. 2020, 10, 14918. [Google Scholar] [CrossRef]
- Poveda, J. Use of plant-defense hormones against pathogen-diseases of postharvest fresh produce. Physiol. Mol. Plant Pathol. 2020, 111, 101521. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Poveda, J. Investigación Básica y Aplicada En La Interacción Trichoderma-Brassicaceae. Ph.D. Thesis, Universidad de Salamanca, Salamanca-España, Spain, 2018. [Google Scholar]
- Livak, K.; Schmittgen, T. Analysis of Relative Gene Expression Data Using RealTime Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kassambara, A. Ggpubr R Package: Ggplot2-Based. Publication Ready Plots. 2019. Available online: http://www.sthda.com/english/articles/24-ggpubr-publication-ready-plots/ (accessed on 31 October 2020).
- McKinnon, S. Package Lemon: Freshing Up Your “Ggplot2” Plots. CRAN. 2020. Available online: https://cran.r-project.org/web/packages/lemon/lemon.pdf (accessed on 31 October 2020).
- Hossain, M.M.; Sultana, F.; Kubota, M.; Koyama, H.; Hyakumachi, M. The plant growth-promoting fungus Penicillium simplicissimum GP17-2 induces resistance in Arabidopsis thaliana by activation of multiple defense signals. Plant Cell Physiol. 2007, 48, 1724–1736. [Google Scholar] [CrossRef] [Green Version]
- de Wit, P. How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef]
- Bailey, B. Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology 1995, 85, 1250–1255. [Google Scholar] [CrossRef]
- Tian, H.; Zhang, Y. The emergence of a mobile signal for systemic acquired resistance. Plant Cell 2019, 31, 1414–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Zhang, Y. Short- and long-distance signaling in plant defense. Plant J. 2021, 105, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Bouizgarne, B.; El-Maarouf-Bouteau, H.; Frankart, C.; Reboutier, D.; Madiona, K.; Pennarun, A.M.; Monestiez, M.; Trouverie, J.; Amiar, Z.; Briand, J.; et al. Early physiological responses of Arabidopsis thaliana cells to fusaric acid: Toxic and signalling effects. New Phytol. 2006, 169, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Betsuyaku, S.; Katou, S.; Takebayashi, Y.; Sakakibara, H.; Nomura, N.; Fukuda, H. Salicylic Acid and Jasmonic Acid Pathways are Activated in Spatially Different Domains around the Infection Site during Effector-Triggered Immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Na, R.; Gijzen, M. Escaping Host Immunity: New Tricks for Plant Pathogens. PLoS Pathog. 2016, 12, e1005631. [Google Scholar] [CrossRef]
- Xin, X.-F.; Kvitko, B.; He, S. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Patel, Z.M.; Mahapatra, R.; Jampala, S.S.M. Role of fungal elicitors in plant defense mechanism. In Molecular Aspects of Plant Beneficial Microbes in Agriculture; Elsevier: Amsterdam, The Netherlands, 2020; pp. 143–158. [Google Scholar]
- Davies, D.R.; Bindschedler, L.V.; Strickland, T.S.; Bolwell, G.P. Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: Implications for basal resistance. J. Exp. Bot. 2006, 57, 1817–1827. [Google Scholar] [CrossRef] [Green Version]
- Osier, T.L.; Lindroth, R.L. Genotype and environment determine allocation to and costs of resistance in quaking aspen. Oecologia 2006, 148, 293–303. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; AL-Harrasi, A. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- de Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Baroja-Fernández, E.; Almagro, G.; Sánchez-López, Á.M.; Bahaji, A.; Gámez-Arcas, S.; de Diego, N.; Dolezal, K.; Muñoz, F.J.; Climent Sanz, E.; Pozueta-Romero, J. Enhanced Yield of Pepper Plants Promoted by Soil Application of Volatiles From Cell-Free Fungal Culture Filtrates Is Associated With Activation of the Beneficial Soil Microbiota. Front. Plant Sci. 2021, 12, 752653. [Google Scholar] [CrossRef] [PubMed]
- García-Gómez, P.; Bahaji, A.; Gámez-Arcas, S.; Muñoz, F.J.; Sánchez-López, Á.M.; Almagro, G.; Baroja-Fernández, E.; Ameztoy, K.; de Diego, N.; Ugena, L.; et al. Volatiles from the fungal phytopathogen Penicillium aurantiogriseum modulate root metabolism and architecture through proteome resetting. Plant Cell Environ. 2020, 43, 2551–2570. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Calderón, E.; Aviles-Garcia, M.E.; Castulo-Rubio, D.Y.; Macías-Rodríguez, L.; Ramírez, V.M.; Santoyo, G.; López-Bucio, J.; Valencia-Cantero, E. Volatile compounds from beneficial or pathogenic bacteria differentially regulate root exudation, transcription of iron transporters, and defense signaling pathways in Sorghum bicolor. Plant Mol. Biol. 2018, 96, 291–304. [Google Scholar] [CrossRef]
- Fincheira, P.; Quiroz, A. Microbial volatiles as plant growth inducers. Microbiol. Res. 2018, 208, 63–75. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).