Identification and Characterization of PHT1 Transporters Family and Differential Expression Patterns in Control and Blindness Broccoli Plants †
Abstract
:1. Introduction
2. Material and Methods
2.1. Identification of Putative Broccoli PHT1 Transporters (PHT1)
2.2. Protein Characterization, Sequence Analysis and Phylogenetic Studies
2.3. RNA-Seq Analysis
2.4. Data Analysis
3. Results
3.1. Genome-Wide Identification of PHT1 Genes in Broccoli and Phylogenetic Analysis
3.2. Chromosomal Location and Protein Features and Subcellular Location Predictions
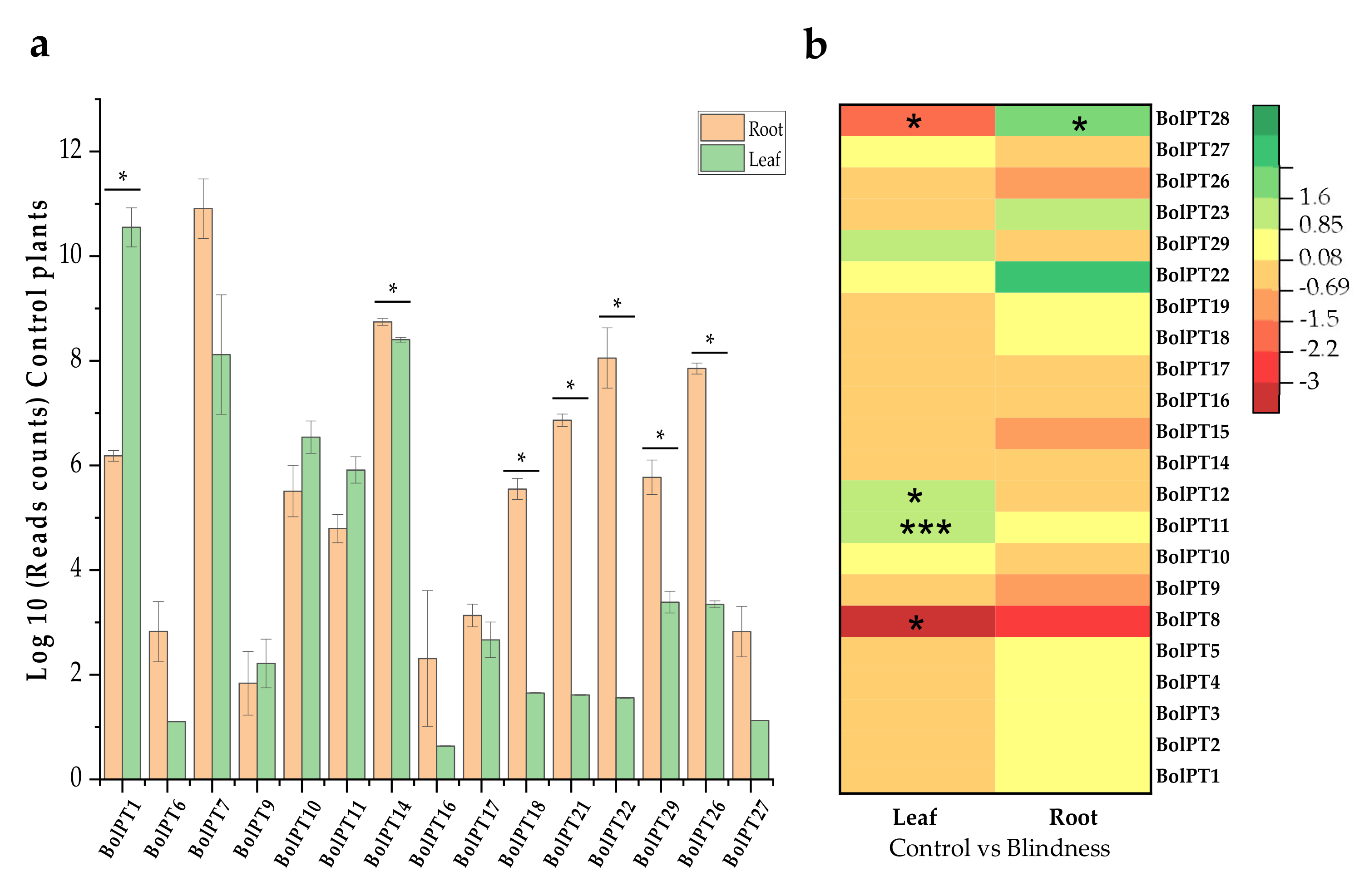
3.3. RNA-Seq Analysis and Expression Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno:, D.A.; Carvajal, M.; López-Berenguer, C.; García-Viguera, C. Chemical and biological characterisation of nutraceutical compounds of broccoli. J. Pharm. Biomed. Anal. 2006, 41, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- El-Bassiony, A.E.M.M.; Shedeed, S.I.; Fawzy, Z.F.; Abd El-Aal, F.S. Influence of different levels of phosphorus supply on growth, yield and quality of some broccoli varieties under sandy soil. Biosci. Res. 2017, 14, 694–704. [Google Scholar]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus Uptake by Plants: From Soil to Cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive genomic identification and expression analysis of the phosphate transporter (PHT) gene family in apple. Front Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussaume, L.; Kanno, S.; Javot, H.; Marin, E.; Nakanishi, T.M.; Thibaud, M.C. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, M.; Chen, A.; Sun, S.; Xu, G. Complex Regulation of Plant Phosphate Transporters and the Gap between Molecular Mechanisms and Practical Application: What Is Missing? Mol. Plant. 2016, 9, 396–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.; Shin, H.-S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Zhang, H.; Wang, S.; Ye, X.; Shi, L.; Xu, F.; Ding, G. Molecular identification of the phosphate transporter family 1 (PHT1) genes and their expression profiles in response to phosphorus deprivation and other abiotic stresses in Brassica napus. PLoS ONE 2019, 14, e0220374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant pan-genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, A.; David, P.; Arrighi, J.F.; Chiarenza, S.; Thibaud, M.C.; Nussaume, L.; Marin, E. Reducing the genetic redundancy of arabidopsis phosphate transporter1 transporters to study phosphate uptake and signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghothama, K.G. Phosphate Acquisition. Annu. Rev. Plant Biol. 2003, 50, 665–693. [Google Scholar] [CrossRef] [PubMed]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]

| Name | Gene ID | Chr loc | No aa | Mw (g/mol) | TMHMM | Cellular Location |
|---|---|---|---|---|---|---|
| BolPT1 | BolC2t09393H | 2 | 538 | 59,106, 69 | 12 | E.R. 1, P.M. 2 |
| BolPT2 | BolC6t40089H | 6 | 537 | 59,136, 52 | 12 | E.R. 1, P.M. 2 |
| BolPT3 | BolC6t38329H | 6 | 551 | 60,907, 72 | 12 | E.R. 1, P.M. 2 |
| BolPT4 | BolC6t40088H | 6 | 538 | 59,064, 55 | 12 | E.R. 1, P.M. 2 |
| BolPT5 | BolC8t49671H | 8 | 557 | 61,462, 94 | 12 | E.R. 1, P.M. 2 |
| BolPT6 | BolC2t10437H | 2 | 517 | 56,464, 16 | 11 | P.M. 1,2 |
| BolPT7 * | BolC9t55444H | 9 | 444 | 48,159, 46 | 10 | P.M. 1,2 |
| BolPT8 | BolC9t55475H | 9 | 509 | 55,452, 65 | 11 | P.M. 2, Vacuole 1 |
| BolPT9 | BolC8t50596H | 8 | 531 | 58,264, 55 | 11 | P.M. 2, Vacuole 1 |
| BolPT10 | BolC4t22465H | 4 | 534 | 58,586, 23 | 11 | P.M. 2, Vacuole 1 |
| BolPT11 | BolC4t28201H | 4 | 529 | 57,910, 35 | 11 | P.M. 2, Vacuole 1 |
| BolPT12 * | BolC4t22466H | 4 | 417 | 46,203, 77 | 8 | P.M. 2, Vacuole 1 |
| BolPT13 | BolC2t10439H | 2 | 521 | 57,251, 2 | 11 | P.M. 2, Vacuole 1 |
| BolPT14 | BolC7t43120H | 7 | 521 | 57,250, 22 | 11 | P.M. 2, Vacuole 1 |
| BolPT15 | BolC9t55490H | 9 | 521 | 57,222, 17 | 11 | P.M. 2, Vacuole 1 |
| BolPT16 | BolC9t55477H | 9 | 521 | 57,220, 24 | 11 | P.M. 2, Vacuole 1 |
| BolPT17 | BolC9t55480H | 9 | 521 | 57,204, 24 | 11 | P.M. 2, Vacuole 1 |
| BolPT18 | BolC2t10445H | 2 | 535 | 58,692, 65 | 11 | P.M. 1,2 |
| BolPT19 | BolC7t43121H | 7 | 535 | 58,392, 07 | 11 | P.M. 1,2 |
| BolPT20 | BolC3t14963H | 3 | 535 | 58,656, 36 | 12 | P.M. 1,2 |
| BolPT21 | BolC4t25357H | 4 | 540 | 59,244, 65 | 11 | P.M. 1,2 |
| BolPT22 | BolC4t22464H | 4 | 533 | 58,692, 05 | 12 | P.M. 1,2 |
| BolPT29 † | BolC4t27979H | 4 | 112 | 11,940, 99 | 1 | P.M. 1,2 |
| BolPT23 | BolC5t30648H | 5 | 542 | 59,950, 3 | 12 | P.M. 1,2 |
| BolPT24 * | BolC7t43115H | 7 | 464 | 50,582, 37 | 11 | P.M. 2, Vacuole 1 |
| BolPT25 * | BolC2t10440H | 2 | 450 | 48,991, 7 | 11 | P.M. 2, Vacuole 1 |
| BolPT26 | BolC6t40092H | 6 | 506 | 56,402, 57 | 12 | P.M. 1,2 |
| BolPT27 | BolC9t55476H | 9 | 521 | 57,381, 33 | 11 | P.M. 1,2 |
| BolPT30 † | BolC9t55487H | 9 | 176 | 19,122, 03 | 4 | P.M. 2, Vacuole 1 |
| BolPT31 † | BolC2t10438H | 2 | 148 | 15,945, 88 | 3 | P.M. 2, Vacuole 1 |
| BolPT28 | BolC3t14547H | 3 | 546 | 59,316, 62 | 10 | P.M. 2, Vacuole 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolas-Espinosa, J.; Carvajal, M. Identification and Characterization of PHT1 Transporters Family and Differential Expression Patterns in Control and Blindness Broccoli Plants. Biol. Life Sci. Forum 2022, 11, 66. https://doi.org/10.3390/IECPS2021-11950
Nicolas-Espinosa J, Carvajal M. Identification and Characterization of PHT1 Transporters Family and Differential Expression Patterns in Control and Blindness Broccoli Plants. Biology and Life Sciences Forum. 2022; 11(1):66. https://doi.org/10.3390/IECPS2021-11950
Chicago/Turabian StyleNicolas-Espinosa, Juan, and Micaela Carvajal. 2022. "Identification and Characterization of PHT1 Transporters Family and Differential Expression Patterns in Control and Blindness Broccoli Plants" Biology and Life Sciences Forum 11, no. 1: 66. https://doi.org/10.3390/IECPS2021-11950
APA StyleNicolas-Espinosa, J., & Carvajal, M. (2022). Identification and Characterization of PHT1 Transporters Family and Differential Expression Patterns in Control and Blindness Broccoli Plants. Biology and Life Sciences Forum, 11(1), 66. https://doi.org/10.3390/IECPS2021-11950







