Abstract
The continuous expansion of freshwater fish culture is confronted with environmental problems owing to their high antibiotic inputs and antibiotic-resistant bacteria. This study explored the antibiotic resistance gene (ARG) profiles of water and fish gut contents from three fishponds in Tianjin using a metagenomics approach. Proteobacteria, Bacteroidetes, and Actinobacteria were the most abundant phyla in all water samples. However, the microbial composition of the fish guts was distinctly different among the three aquaculture farms. Cetobacterium, Bacillus, Weissella, and Fusobacterium were the dominant genera in the gut contents of all fish. More than 20 unique ARGs with relatively high abundances were detected in both water and fish gut content samples. The dominant genes and pathways of antibiotic resistance mechanisms detected in all samples were antibiotic efflux, antibiotic inactivation, antibiotic target alteration, antibiotic target protection, antibiotic target replacement, and reduced permeability to antibiotics. In addition, our results indicate that antibiotics, such as florfenicol, and heavy metals, such as Zn and Cu, could have a significant correlation with some common ARGs, indicating that antibiotic-resistant bacteria could co-occur with heavy metals. Our study provides a research basis for the development of a strategy for practical antibiotic application and heavy metal monitoring in aquaculture.
1. Introduction
Antibiotic resistance has become an increasing global problem that threatens effective antibiotic therapies and the healthcare system. ARGs are found in many environments and are regarded as emerging contaminants of global concern [1]. Long-term overuse of antibiotics in aquaculture has caused propagation and acquisition of drug resistance [2]. However, fish consumption makes up an important part of the human diet around the world and provides various important nutrients that are essential to good health [3]. China (mainland) produced more than 64.13% (49,620.1 thousand tons) of total aquaculture animal production in the world [4], and Tianjin province produced about 237.97 thousand tons of total aquaculture products in China [5]. Pond culture is the primary form of aquaculture in China, which accounts for 48.84% of the total production of aquatic products [6]. However, the aquaculture systems have been reported to led to the spread of ARGs and antibiotic-resistant bacteria [2]. According to previous studies, exposure to contaminants such as antibiotics and metals may promote the spread of antibiotic resistance in the environment [7,8,9,10]. An increasing number of researchers have started to pay attention to the influences of antibiotics and metals on ARGs [8,9,10].
Metagenomic analysis is a culture-independent molecular approach that has provided powerful tools to explore the dissemination of ARGs in different environments [11,12]. The metagenomic approach does not depend on specific primers which are designed from known target sequences, nor does it depend on the detection of unknown ARGs. With this untargeted analysis method, more information on the prevalence and diversity of ARGs can be recovered, thereby making it possible to discover novel ARGs [13]. To date, the metagenomics method has been used to characterize the existence of ARGs in a variety of environments, including salt lakes, freshwater, sharpbelly (Hemiculter leucisculus), and drinking water [14,15,16,17]. However, few reports are available on the investigation of ARGs in fish gut contents and their aquaculture environments using a metagenomics-based approach.
Tianjin plays a vital role in the aquaculture industry in northern China owing to its geological advantages. Here, we report the bacterial community composition and diversity of ARGs in three aquaculture farms in Tianjin. We identified the relevant high-risk factors, such as antibiotics and metals, and carried out a correlation analysis between these high-risk factors and ARGs. The results of this study will help us to gain a deeper understanding of the bacterial communities and ARGs in pond aquaculture environments. Moreover, they exhibited the co-occurrence network between ARGs and other factors, including bacterial communities, antibiotics, and heavy metals. This study showed that aquaculture system was important ARGs reservoirs.
2. Materials and Methods
2.1. Sample Collection
All samples were collected from three aquaculture farms (LK, BXL, and LZG) in the Baodi District of Tianjin, China. The fish in the LK farm were largemouth bass (Micropterus salmoides), which originated from North America. Fish in BXL and LZG farms were grass carp (Ctenopharyngodon idella) and common carp (Cyprinus carpio), respectively. Ten fish were collected from each farm, followed by the delivery to the laboratory at 4 ℃. After aseptically opening the abdominal cavity, fish gut contents from three farms were gently squeezed out and polled into 15 mL sterile centrifuge tubes, respectively. Then every mixture was divided into three portions and stored at −80℃. In this study, each kind of cultured adult fish were in similar size (largemouth bass: 30.22 ± 0.57 cm length and 586.55 ± 40.44 g fresh weight; grass carp: 35.78 ± 0.82 cm length and 748.42 ± 65.34 g fresh weight; common carp: 36.49 ± 0.73 cm length and 929.76 ± 84.57 g fresh weight).
Water samples from each farm were collected using sterile brown bottles prewashed more than three times with water from the sampling locations. A 5 L plexiglass water bucket was used to collect water for sampling. All water samples were immediately transported to the laboratory at 4 °C for further analysis.
2.2. Antibiotics and Metals Quantification
The antibiotics analyzed in this study included danofloxacin mesylate, sulfamonomethoxine, sulfadimethoxine, trimethoprim, sulfamethizole, sulfamerazine, sulfadiazine, sulfisoxazole, sulfamethoxydiazine, florfenicol, roxithromycin, orbifloxacin, fleroxacin, azithromycin, lincomycin, enoxacin, difloxacin, enrofloxacin, lomefloxacin, oxytetracycline, doxycycline, timicosin, ciprofloxacin, norfloxacin, flumequine, and ofloxacin. For pretreatment of fish gut content, samples (~1 g) were successively extracted with acetonitrile and acidified acetonitrile. The extract was then purified using a fast veterinary drug extraction kit (FaVEx-NM50, Getech, Taiwan). The water samples collected from each farm were successively passed through 0.45 µm and 0.22 µm glass fiber filters to remove suspended solids. Subsequently, the filtered water (~500 mL) was adjusted to pH4 using concentrated HCl (1 V%:1 V%) with water. Next, 1 g of disodium EDTA was added to the filtered water sample and concentrated with an Oasis HLB cartridge (6 mL, Waters, MA, USA). The extracted samples were evaporated using a nitrogen concentrator and re-dissolved in acetonitrile mixed with 0.1% formic acid aqueous solution (v/v, 1:19). Finally, the extracts were filtered through a 0.22 µm organic filter and investigated using HPLC-MS/MS (Xevo TQ-S, ACQUITY UPLC, Waters) according to the method reported by Ding et al. [17]. Pretreatment and antibiotic determination of each sample were conducted in triplicate.
Metals and minerals (V, Cr, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Ag, Cd, Pb, U, and Hg) were analyzed using inductively coupled plasma-mass spectrometry (ICP-MS, Agilent 7900, USA) because they could persist and propagate in aquaculture environments related with ARGs and are commonly detected [18]. Details about the pretreatment methods of samples, various instrument parameters, and the quality control were performed as previously reported [19,20]. All measurements were conducted in triplicate.
2.3. DNA Extraction, Library Preparation and Metagenomic Sequencing
For DNA extraction and sequencing, intestinal content samples of fish and filtered water samples were shipped on dry ice to Novegene (Beijing, China). The potential contamination and degree of degradation of the extracted DNA were assessed using 1% agarose gels. A NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA) was used to determine DNA purity (OD260/OD280 and OD260/OD230). The QubitA® dsDNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, Waltham, MA, USA) was used to measure the DNA concentration. The sequencing library was constructed using DNA contents above 1 μg OD values ranged from 1.8 to 2.0.
DNA fragment preparation was carried out with a total of 1 µg mass of DNA. To associate sequences with specific samples, the NEBNext® UltraTM DNA Library Prep Kit (NEB, Ipswich, MA, USA) for Illumina was used to prepare sequencing libraries. Once the DNA sample had been sonicated to 350 bp, it was end-polished, A-tailed, and ligated with a full-length adaptor for Illumina sequencing. In addition, libraries were analyzed with an Agilent2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) to determine their sizes, followed by real-time PCR quantification (AMPure XP system; Beckman Coulter, Fullerton, CA, USA).
Based on the manufacturer’s recommendations, cBot Cluster Generation System was used to cluster the index-coded samples. On an Illumina HiSeq platform (Illumina, San Diego, CA, USA), paired-end reads were generated from the library preparations. Readfq (V8) was used to remove raw reads obtained from the Illumina HiSeq sequencing platform that contained >40 bp sequences with a quality value less than 38, overlapped more than 15 bp with adapter sequences, or had an N base run longer than 10 bp. After quality filtering, clean reads were BLAST against the host database using Bowtie software (V2.2.4) to filter the reads that were of host origin (largemouth bass Micropterus salmoides, common carp Cyprinus carpio, grass carp Ctenopharyngodon idellus). Clear reads of samples taken from the fish intestine content were assembled into scaftigs using SOAPdenovo software (V2.04). For the samples taken from aquaculture farms, the MEGAHIT assembler (V1.04-beta) using the options --presets meta-large, -min-count 2, -k min 27, -k-max 87, -k-step 10 [21] to assemble the clean data. After the assembly of the metagenome, the assembled scaftigs were interrupted at the N connection and the scaftigs without an N were maintained [22,23,24]. The clean data of all samples were compared to each scaffold using Bowtie 2.2.4 software to acquire the PE reads. Assembling the unused reads from each sample was performed using MEGAHIT with the same parameters for each sample. Scaftigs generated from both single and mixed assemblies that were longer than 500 bp were used for statistical analysis.
2.4. Gene Prediction from Scaftigs
Open reading frames (ORFs) present in the scaftigs (≥500 bp) assembled from both single and mixed sequences were determined using MetaGeneMark (V2.10) software, and ORFs with a length shorter than 100 bp were filtered out [23,24,25,26,27] using default parameters. A non-redundant gene catalogue was then constructed using CD-HIT (V4.5.8) [28,29]. The previously reported parameters options of -c 0.95, -G 0, -a 0.9, -g 1, -d 0 were used [27,30]. To determine the abundance of genes, the reads were mapped to the initial gene catalogue using Bowtie2.2.4 using the options --end-to-end, --sensitive, -I 200, -X 400 [19,20]. For further analysis, only genes with at least two mapped reads were considered unigenes [25,31]. In order to calculate gene abundance, the number of reads was counted, and the length of the genes was normalized [31,32].
2.5. Taxonomic Classification
DIAMOND (V0.9.9) with the options -blastp and -e 1e-5 [33] was used to search each sample’s unigenes against the NCBI NR database (Version: 2 January 2018) with an E-value cut-off of <1 × 10 − 20. Using MEGAN’s lowest common ancestor-based approach, each gene’s taxonomic rank was determined [34]. By adding the abundance of the genes annotated to a feature, the abundance of a taxonomic group was estimated.
2.6. ARGs Analysis
Using the Resistance Gene Identifier (RGI), unigenes were searched for ARGs against the widely accepted ARG database, CARD (V2.0.1) [35,36,37]. Based on the alignment results, the relative abundance of ARG-like sequences was obtained. In “total ARG-like sequences” and “total metagenome sequences,” the percentages of various ARG-like sequence types were defined as “percentage” (%) and “abundance” (ppm, one read in 1 million reads), respectively [38,39].
3. Results
3.1. Bacterial Community Composition
The relative abundances of bacterial communities at the phylum level in the different samples are shown in Figure 1a. The phylum level composition of the bacterial community revealed Proteobacteria as the predominant bacteria (16%, 20%, and 24% on average, respectively) in all water samples collected from three different fish farms (LK, BXL, and LZG). Bacteroidetes and Actinobacteria were the two other most abundant phyla in the water samples. Additionally, Planctomycetes was dominant in the BXL and LZG water samples, with percentages of 9% and 4%, respectively. However, the fish gut content samples showed a distinct microbial community composition (Figures S2 and S3). The predominant phylum in fish gut content samples collected from LK was Firmicutes, with a mean abundance of 58%, followed by Fusobacteria (8%) and Proteobacteria (8%). Fusobacteria and Proteobacteria were the two dominant phyla among the fish gut content samples collected from BXL (40% and 5% on average, respectively) and LZG (4% and 3% on average, respectively) farms.
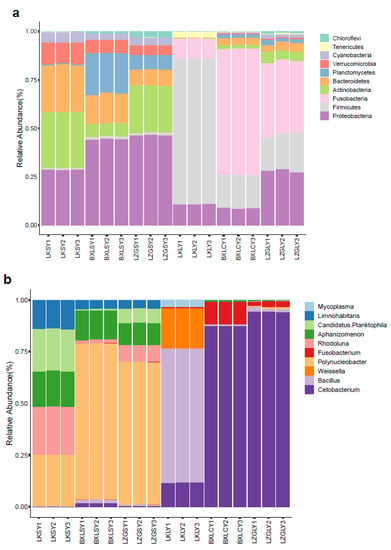
Figure 1.
The profile of bacterial communities in different samples. (a) phylum level; (b) genus level. LK, BXL, and LZG were the abbreviation of the three fish farms. Water samples from LK farm were LKSY1, LKSY2 and LKSY3 while LKLY1, LKLY2 and LKLY3 were the name of fish gut content samples from LK fish farm. Likewise, BXLSY1-3 and LZGSY1-3 were water samples from BXL and LZG fish farm, respectively. BXLCY1-3 and LZGLY 1-3 were fish gut contents from BXL and LZG fish farm, respectively.
Microbial community structures also varied among the samples at the genus level (Figure 1b and Figure S3). Polynucleobacter, Rhodoluna, and Candidatus Planktophila were the predominant genera in all water samples, except samples from BXL farms, in which only Polynucleobacter was relatively abundant. Bacillus, Weissella, and Cetobacterium were abundant in the fish gut content samples which were collected from LK farms, with percentages of 42%, 10%, and 4%, respectively. Cetobacterium and Fusobacterium were the two dominant genera among fish gut content samples from the BXL farm, whereas only Cetobacterium was dominant among samples from the LZG farm. All of the above data showed different microbial community compositions in different aquaculture environments. In particular, bacterial community diversity was more complex in fish gut content samples. In the current study, different bacterial community structures were reported in different types of fish guts. These complex bacterial community compositions play vital roles in the distribution of ARGs. A heatmap of the bacterial composition diversities shows detailed differences at the phylum level (Figure 2).
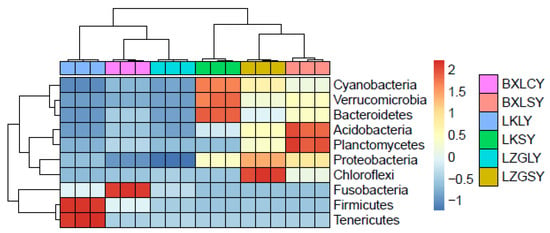
Figure 2.
Heatmap of bacterial composition diversities in different samples (phylum level).
BXLCY, LKLY and LZGLY were fish gut content sample groups of BXL, LK and LZG fish farm, respectively. All water sample groups were named as LKSY, BXLSY and LZGSY.
3.2. Diversity and Abundance of ARGs
As shown in Figure 3, 20 unique ARGs (aac(6′)-I30, otr(A), dfrA26, lnu(C), lnu(B), CfiA(CcrA), blaGIM-2, tet(W/N/W), tetB(P), optrA, aac(2′)-IIb, vatI, adeF, vanX-O, blaTEM-187, blaTEM-67, sul1, catA16(catQ), farB, and ant(9)-Ia) with relatively higher abundance were detected in all samples. The detected frequency of higher abundance level genes implied that samples from fish guts (LKLY, BXLCY, and LZGLY) were associated with the diversity of ARGs. The high relative abundance of ARGs in all samples conferred resistance to tetracycline, aminoglycosides, quinolones, and phenicols, might imply extensive use of these common antibiotics in these aquaculture systems. Additionally, the occurrence of β-lactamase resistance genes implied that clonal transmission has occurred between bacterial genera in aquaculture and human medical practice. As reported in a previous study [40], livestock might form a reservoir for extended-spectrum beta-lactamase (ESBL)- and AmpC beta-lactamase-encoding resistance genes, capable of being transmitted to humans. This risk should be pursued in future studies on the vital status of ESBL/AmpC beta-lactamase antibiotics. Notably, vatI (streptogramin resistance gene), vanX-O (vancomycin resistance gene), and adeF (multidrug resistance gene) were detected in some fish gut content samples. The fish could be risk factors for the transmission of antibiotic resistance genes, such as vatI, vanX-O, and adeF, to humans. Transmission via the food chain has also been suggested [41,42]. A heatmap of ARGs shows detailed differences at the phylum level (Figure 4).
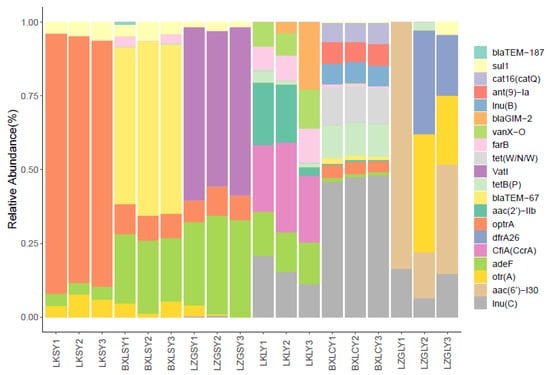
Figure 3.
Relative abundances of ARGs in different samples. LK, BXL, and LZG were the abbreviation of the three fish farms. Water samples from LK farm were LKSY1, LKSY2, and LKSY3 while LKLY1, LKLY2, and LKLY3 were the name of fish gut content samples from LK fish farm. Likewise, BXLSY1-3 and LZGSY1-3 were water samples from BXL and LZG fish farm, respectively. BXLCY1-3 and LZGLY 1-3 were fish gut contents from BXL and LZG fish farm, respectively.
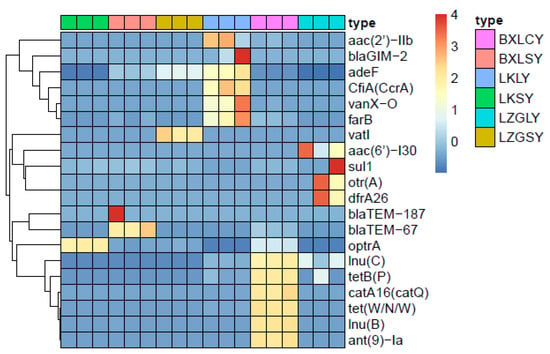
Figure 4.
Heatmap of ARGs in different samples. BXLCY, LKLY and LZGLY were fish gut content sample groups of BXL, LK and LZG fish farm, respectively. All water sample groups were named as LKSY, BXLSY and LZGSY.
Analysis of the diversity and abundance of ARGs showed that otrA, optrA, adeF, and sul1 were the predominant ARGs in all water samples collected from three different fish pounds (LK, BXL, and LZG), implying that common antibiotics such as tetracycline, florfenicol, quinolone, and sulfonamide were used intensively. In addition, the antibiotic resistance genes blaTEM-67 (beta-lactam resistance gene) and vatI (streptogramin resistance gene) were detected in water samples from BXL and LZG farms, respectively. However, beta-lactam antibiotics are not common in aquaculture systems. It was concluded that these two aquaculture environments are greatly affected by human activities. In all the fish gut content samples (LKLY, BXLCY, and LZGLY), lnuC was detected at a relatively high abundance. Furthermore, lnuB, which belongs to the same antimicrobial resistance (AMR) gene family as lnuC, was found in the BXLCY samples. The aminoglycoside antibiotic resistance gene aac(6′)-I30, which is highly abundant, only occurred in fish gut content samples collected from LZG farms. Different ARG compositions of the fish gut contents can be explained by the use of antibiotics in part among different aquaculture environments.
3.3. Antibiotic Resistance Mechanisms of the ARGs
To evaluate the potential risk to public health, it is very important to elucidate the antibiotic resistance mechanisms of the ARGs prevalent in these aquaculture environments. Many antibiotic resistance mechanisms are strongly related to microbial metabolic activity [43].
As shown in Figure 5, antibiotic resistance mechanisms are distributed among different phyla. The dominant types of antibiotic resistance mechanisms reported in all samples were antibiotic efflux, antibiotic inactivation, antibiotic target alteration, antibiotic target protection, and antibiotic target replacement. Proteobacteria were the dominant bacteria in these three aquaculture systems with antibiotic resistance mechanisms characterized by antibiotic efflux, antibiotic inactivation, and antibiotic target alteration. This was the same for Actinobacteria and Bacteroidetes. Notably, it has been reported that most drug efflux pumps confer a multidrug resistance phenotype to a large variety of substrates, including several classes of antibiotics and non-antibiotic drugs [44]. Currently, efflux pumps have been identified as fluoroquinolone- and tetracycline-resistance mechanisms. Penicillinase and aminoglycoside-modification enzymes are typical enzymes involved in antibiotic inactivation, whereas antibiotic resistance mechanisms of antibiotic target alteration have been reported in methicillin-resistant strains and vancomycin-resistant strains [45].
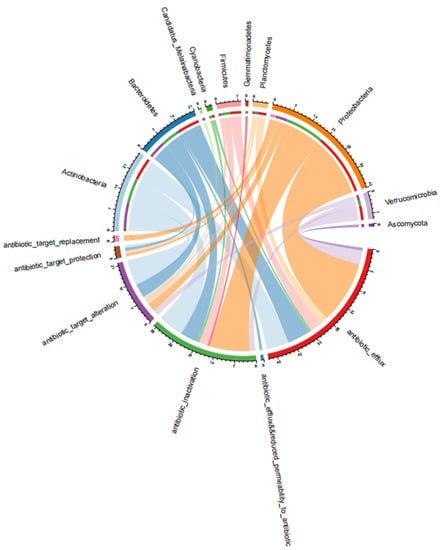
Figure 5.
The relationships between antibiotic resistance mechanisms and bacteria phyla. Resistance mechanisms and bacteria phyla were represented by different colors in the circle.
3.4. Relationship between ARGs and Microbial Community
The existence of ARGs is associated with the composition diversities of bacterial communities. Bacteria harboring different ARGs may be affected by different environmental conditions, resulting in a change in the ARG profiles of different samples. The correlation between the bacterial communities and ARGs profiles could be investigated using a metagenomic analysis approach. Proteobacteria, Actinobacteria, and Bacteroidetes were the three dominant phyla in antibiotic-resistant bacteria among the water samples collected from the three different fish farms (Figure 6). Especially for Actinobacteria and Verrucomicrobia in all water samples, the proportion of antibiotic-resistant bacteria was much greater than that in the total microbial communities, indicating that Actinobacteria and Verrucomicrobia were more resistant than other phyla.
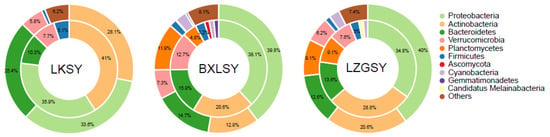
Figure 6.
Attribution analysis of ARGs and bacteria phyla in all water samples showed by circle map. The distribution of bacteria phyla of the total ARGs was represented in inner circle, and the phyla distribution of all samples was in the outer circle.
Notably, in all fish gut content sample groups, the proportion of the most dominant phyla in the antibiotic-resistant bacteria was greater than that of the total microbial community (Figure S1). Proteobacteria was the most dominant phylum among antibiotic-resistant bacteria in LKLY and BXLCY, with proportions of 66.7% and 37.4%, respectively. Firmicutes was the second dominant phylum among antibiotic-resistant bacteria in these two groups (proportions of 33.3% and 18.8%, respectively), whereas it was the most dominant phylum in LZGLY (proportion of 100%). It was concluded that Proteobacteria and Firmicutes carried more resistance genes than other phyla in the fish gut contents.
3.5. Relationship between ARGs and Antibiotics
Spearman’s correction analysis was employed to explore the potential antibiotic factors responsible for the differences in the ARG profiles among the samples (Figure 7).
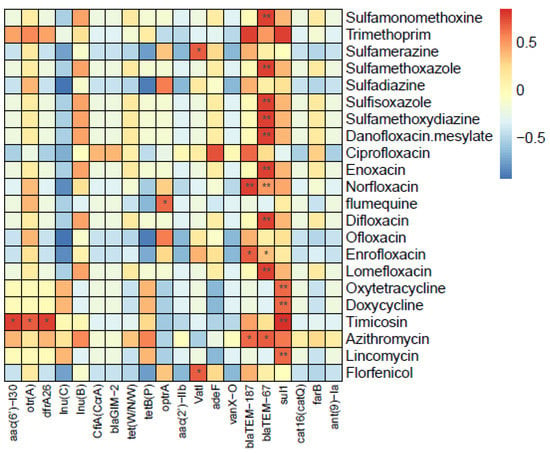
Figure 7.
Heatmap of relative abundance of ARGs with antibiotics. *: significant difference, p < 0.05; **: highly significant difference, p < 0.01.
The 20 most abundant ARGs were selected for detailed analysis of the correlations between antibiotics and antibiotic resistance genes. Among the ARGs selected, only 8 ARGs showed significant correlation with common antibiotics detected in this study. Eleven antibiotics showed significant positive correlations with blaTEM-67, which was associated with the highest number of antibiotics in this study. The following was sul1, which was significantly positively correlated to four antibiotics, such as oxytetracycline, doxycycline, timicosin, and lincomycin. Another beta-lactam antibiotic resistance gene, blaTEM-187, showed a significant positive correlation with norfloxacin, enrofloxacin, and azithromycin. Three different types of ARGs, aac(6′)-I30, otr(A), and dfrA26, were strongly correlated with timicosin. In addition, optrA, a phenicol (florfenicol) antibiotic resistance gene, was found to be significantly associated with flumequine, and VatI, which is a streptogramin antibiotic resistance gene, showed a significant positive correlation with florfenicol. The antibiotics concentrations were presented in Tables S1 and S2.
In addition, some antibiotics such as doxycycline and sulfadiazine also correlate with some heavy metals. Sulfadiazine showed striking negative associations with the heavy metals Ag, Se, and Cu. In contrast, doxycycline, oxytetracycline, and lincomycin levels were positively correlated with Zn levels. More details regarding the ARGs and different environmental factors are presented in Figure 8.
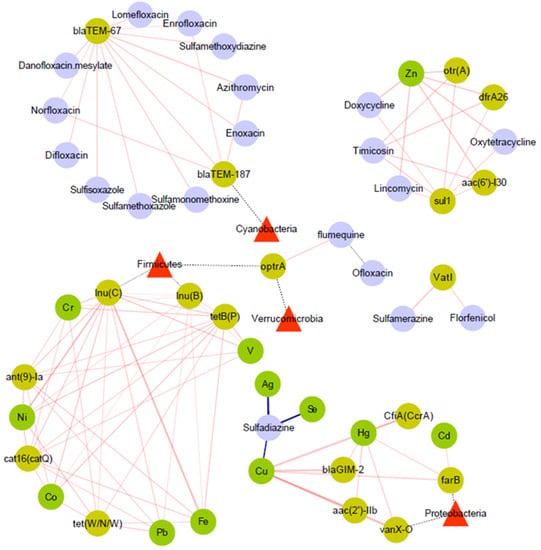
Figure 8.
Network analysis of correlation. Margins are weighted according to the correlation coefficient. Red margins stand for positive correlation, blue margins stand for negative correlation, dash lines only mean the attribution of ARGs from different bacteria in phylum level. There is only significant correlation represented (p < 0.05).
3.6. Associations between ARGs and Heavy Metals
Significant correlations between heavy metals and ARGs were detected in the samples, as shown in the heatmap, delineating the significant impact of heavy metals on ARGs (Figure 9). Some common ARGs, such as lnu(C), lnu(B), tet(W/NW), tetB(P), cat16(catQ), and ant(9)-Ia, were influenced by many kinds of heavy metals. In our investigation, V, Cr, Fe, Co, Ni, Cu, Pb, and Hg were the main heavy metals positively correlated with many kinds of ARGs, including beta-lactam, tetracycline, phenicol (florfenicol), and aminoglycoside antibiotic resistance genes. Notably, otr(A) was significantly negatively associated with Zn. However, the impact of these negatively correlated heavy metals should not be ignored. Generally, metal pollution may significantly affect the maintenance of a broad spectrum of ARGs. A considerable effort may be required to mitigate this risk by studying metal-induced co-selection of antibiotic resistance genes in metal-rich environments and the potential implications of this on public health [46]. The concentration of heavy metals could be found in Tables S3 and S4.
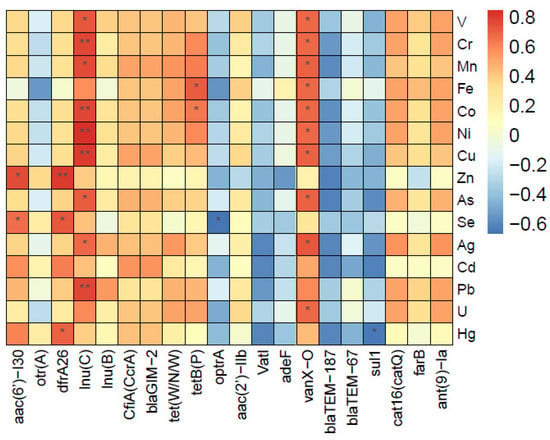
Figure 9.
Heatmap of relative abundance of ARGs with heavy metals. *: significant difference, p < 0.05; **: highly significant difference, p < 0.01.
More details regarding heavy metals, ARGs, antibiotics, and some main phyla are presented in Figure 8. As shown in Figure 8, heavy metals play an important role in the distribution and propagation of ARGs and antibiotics. Likewise, the distribution of heavy metals and antibiotics also plays a major role in the shaping of bacterial communities.
4. Discussion
In addition to affecting the human sector, antibiotic resistance affects the food industry and the environment. In a recent EFSA scientific opinion [47], the EFSA recommends further investigation of ARGs in food and food-producing environments, especially aquaculture environments. Only a few studies have examined microbial communities and ARGs in fish gut contents and the relevant aquaculture environments. The presence and distribution of ARGs are closely associated with microbial structures and other relevant environmental factors in aquaculture. In this study, a metagenomic sequencing approach was used to simultaneously analyze and detect bacterial diversity and hundreds of ARGs. We profiled the bacterial community and antibiotic resistance genes in three types of fish gut contents and their aquaculture environments. The correlation between ARGs and other factors such as microbial diversity, antibiotics, and heavy metals was analyzed.
Proteobacteria, Actinobacteria, and Bacteroidetes were the main phyla found in all water samples, similarly to other reports on freshwater aquaculture environments [40,48]. According to previous studies, many antibiotics can be resistant to Proteobacteria and Bacteroidetes, including tetracycline, beta-lactams, aminoglycosides, and erythromycin [41,42]. Relevant ARGs such as otr(A), adeF, and blaTEM-67 were detected in the three water sample groups. However, there were some differences in the relative abundances of these phyla and ARGs in water from different farms. For example, Planctomycetes was the dominant phylum in BXL and LZG water samples (BXLSY and LZGSY), and Chloroflexi were also found to be more predominant in water samples from these two farms than in LK. Notably, Planctomycetes and Chloroflexi were found to be involved in the degradation of organic matter, indicating the organic matter contamination of these two farms. The trends in bacterial communities were similar to the patterns of ARG profiles because the environmental factors, like antibiotics and heavy metals concentration that affect ARGs also influence microbial communities [49].
Proteobacteria, Firmicutes, and Fusobacteria were found to be the three dominant phyla in all fish gut content samples, which is agreement with previous reports [50,51,52]. However, the relative abundances of these phyla in the different fish gut contents were different. In general, fish gut content samples from BXLCY and LZGLY (BXL and LZG) farms shared similar bacterial communities. Planctomycetes and Chloroflexi were also detected in both sample groups. Planctomycetes have been identified as a promising source of bioactive natural products, such as novel antibiotics [53]. Therefore, more attention should be paid to isolating planctomycetes from aquaculture samples in the future. In addition, the significant difference in bacterial communities between fish gut content samples from LK farms (LKLY) and the other two farms resulted from the different proportion of Firmicutes (at the phylum level) or Bacillus (at the genus level). This obvious difference might be a consequence of probiotic application in aquaculture. According to studies, Bacillus species are beneficial probiotics because they produce antimicrobial substances that are active against a wide range of bacteria and are nonpathogenic and nontoxic to fish. In addition, the application of Bacillus species can improve the water quality [54]. This speculation was also in accordance with the lower ARG diversity in water samples from LK farms. Probiotic utilization can reduce the application of antibiotics to decrease the distribution of ARGs to some extent.
The abundance and diversity of ARGs in the fish gut content samples were higher than those in water samples. There were only four ARGs, otr(A), adeF, optrA, and sul1, with relatively higher abundances detected in the LKSY water sample group, all of which existed in other water sample groups as well. Notably, the optrA gene, encoding an ATP-binding cassette F (ABC-F) protein that confers resistance to oxazolidinones and phenicols, was found to be the predominant ARG in the LKSY waste sample group. In previous studies, the presence of optrA was limited to gram-positive bacteria, particularly the genera Enterococcus and Staphylococcus. However, a recent study also detected optrA in some gram-negative bacteria [55]. Therefore, subsequent investigations should closely monitor its presence in of both gram-positive and gram-negative bacteria to avoid dissemination of this novel oxazolidinone and phenicol resistance genes in different kinds of food and the relevant environment. Bacteria containing adeF can simultaneously resist tetracycline and fluoroquinolones. AdeF is carried simultaneously by integrons, transposons, and plasmids in different hosts. Furthermore, it might be distributed and spread extensively through food chains and different environments owing to frequent horizontal gene transfer [56]. Compared to the lower ARG diversity in the water sample groups, the ARG diversity in the three fish gut content samples were much higher. The relatively higher diversity of ARGs in fish gut content samples may be closely associated with more complicated microbial composition structures. In all samples, the dominant antibiotic resistance mechanisms were antibiotic efflux, antibiotic inactivation, antibiotic target alteration, antibiotic target protection, antibiotic target replacement, and reduced permeability to antibiotics. Among all the mechanisms, drug efflux pumps should draw more attention because they confer a multidrug resistance phenotype to a large variety of substrates, including several classes of antibiotics and non-antibiotic drugs [44].
The propagation and stability of ARGs in aquaculture environments are influenced not only by microbial communities, but also by abiotic factors, including organic matter, antibiotics, heavy metals, and so on [57,58]. In this study, we investigated the correlations between antibiotics and heavy metals. Only a few of the 22 selected antibiotics were detected in water or fish gut content samples. The persistence of antibiotics in the environment varies, which ranges from a few hours to even a few months [59]. Therefore, it is possible that the antibiotics had mostly been degraded or biotransformed in these samples. Meanwhile, a plenty of ARGs with various diversities were discovered in all samples, as well as in the samples with no antibiotics detected, indicating that the occurrence of antibiotics was not a precondition for persistence of ARGs. Similarly, several studies have found ARGs, but no relevant antibiotics [60,61]. In the future, more attention should be focused on the mechanisms underlying the persistence of ARGs without external selection pressure. This could contribute to further problems in the distribution of antibiotic resistance. Heavy metals may lead to a shift in the bacterial communities. Significant positive correlations have been reported between microbial communities and heavy metals [62]. Heavy metals could promote the conjugative transfer of ARGs between microbial communities and might enhance the co-selection of ARGs and metal resistance genes under high heavy-metal pressure [63,64]. However, it was notable that some heavy metals were negatively correlated with some ARGs, implying that the heavy metal-selective pressure at low concentrations on ARGs should be drastically investigated. It has also been reported that factors such as some heavy metals affect the propagation of ARGs not only by themselves but also by affecting the bacterial diversities [65].
5. Conclusions
In summary, this study profiled the bacterial communities and ARGs in three different aquaculture environments, including water and fish gut content samples. The dominant bacterial phyla were found to be Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Fusobacteria, and the most common ARGs were adeF, otr(A), and lnu(C) in most of the water and fish gut content samples. The occurrence of ARGs was closely associated with the structure and composition of bacterial communities. In addition, factors including antibiotics and heavy metals play important roles in the distribution of ARGs. Further studies should be conducted to reveal the mechanisms underlying the propagation and persistence of ARGs in different aquaculture environments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/aquacj2040016/s1, Figure S1: Attribution analysis of ARGs and bacteria phyla in all fish gut content samples showed by circle map. Figure S2: Alpha diversity of bacteria community in phylum level. Figure S3: Alpha diversity of bacteria community in genus level. Table S1 Antibiotics concentrations in water samples from three different fish farms. Table S2 Antibiotics concentrations in fish gut content samples from three different fish farms. Table S3 Heavy metals concentrations in water samples from three different fish farms. Table S4 Heavy metals concentrations in fish gut content samples from three different fish farms.
Author Contributions
Conceptualization, Q.W. and X.Z.; methodology, Q.W., Q.D. and Z.W.; software, X.Z.; formal analysis, J.D.; investigation, Y.L., J.Z. and L.Y.; resources, B.C.; data curation, L.G.; writing—original draft preparation, Q.W.; writing—review and editing, X.Z.; visualization, X.Z.; supervision, L.G.; project administration, Q.W.; funding acquisition, P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the BJAST Budding Talent Program of Beijing Academy of Science and Technology, grant number 11000022T000000455559, and the Municipal Financial Project of Beijing Academy of Science and Technology, grant number 11000022T000000442954.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Lu, J.; Zhang, Y.-X.; Wu, J.; Luo, Y.; Liu, H. Metagenomic analysis of antibiotic resistance genes in coastal industrial mariculture systems. Bioresour. Technol. 2018, 253, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Okyere, A.; Bishoff, D.; Oyaro, M.O.; Ajami, N.J.; Darkoh, C. Analysis of Fish Commonly Sold in Local Supermarkets Reveals the Presence of Pathogenic and Multidrug-Resistant Bacterial Communities. Microbiol. Insights 2018, 11, 1178636118786925. [Google Scholar] [CrossRef]
- The State of World Fisheries and Aquaculture (SOFIA) 2022. Available online: http://www.lmcwater.org.cn/authoritative_opinion/study/202207/t20220701_35915.html (accessed on 1 July 2022).
- Fishery Bureau of Ministry of Agriculture. 2022 China Fishery Statistics Yearbook; China Agriculture Press: Beijing, China, 2022.
- Liu, Q.; Lai, Z.; Gao, Y.; Wang, C.; Zeng, Y.; Liu, E.; Mai, Y.; Yang, W.; Li, H. Connection between the Gut Microbiota of Largemouth Bass (Micropterus salmoides) and Microbiota of the Pond Culture Environment. Microorganisms 2021, 9, 1770. [Google Scholar] [CrossRef]
- Lloyd, N.A.; Nazaret, S.; Barkay, T. Whole genome sequences to assess the link between antibiotic and metal resistance in three coastal marine bacteria isolated from the mummichog gastrointestinal tract. Mar. Pollut. Bull. 2018, 135, 514–520. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, Y.; Sun, C.; Wang, W.; Wu, Y.; Fan, L.; Liu, B. Profile of Bacterial Community and Antibiotic Resistance Genes in Typical Vegetable Greenhouse Soil. Int. J. Environ. Res. Public Health 2022, 19, 7742. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Eckert, E.M.; D’Urso, S.; Bertoni, R.; Gillan, D.C.; Wattiez, R.; Corno, G. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 2016, 94, 208–214. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water. Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Zhao, Y.; Li, B.; Huang, C.-L.; Zhang, S.-Y.; Si-Yu, Z.; Chen, Y.-S.; Zhang, T.; Gillings, M.; Su, J.-Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 16270. [Google Scholar] [CrossRef]
- Di Cesare, F. Functional Metagenomics for Identification of Antibiotic Resistance Genes (ARGs). Methods. Mol. Biol. 2021, 2242, 173–183. [Google Scholar]
- Hu, Y.; Jiang, L.; Sun, X.; Wu, J.; Ma, L.; Zhou, Y.; Lin, K.; Luo, Y.; Cui, C. Risk assessment of antibiotic resistance genes in the drinking water system. Sci. Total Environ. 2021, 800, 149650. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Wang, F.; Mu, R.; Huang, J.; Zhao, R.; Li, X.; Yu, K.; Li, B. Metagenomics analysis revealing the occurrence of antibiotic resistome in salt lakes. Sci. Total Environ. 2021, 790, 148262. [Google Scholar] [CrossRef] [PubMed]
- Nnadozie, C.F.; Odume, O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019, 254, 113067. [Google Scholar] [CrossRef]
- Ding, Q.; Wang, J.F.; Feng, Y.C.; Wu, Z.; Ma, L.L.; Liu, J.; Wang, Y.; Jia, L.; Gao, L.J.; Shao, P.; et al. Determination of 32 kinds of antibiotic residues in fish intestinal content by high performance liquid chromatography-tandem mass spectrometry. J. Food Saf. Qual. 2022, 13, 1141–1149. [Google Scholar]
- Xue, X.; Jia, J.; Yue, X.; Guan, Y.; Zhu, L.; Wang, Z. River contamination shapes the microbiome and antibiotic resistance in sharpbelly (Hemiculter leucisculus). Environ. Pollut. 2021, 268, 115796. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Zhao, H. Propagation of antibiotic resistance genes in an industrial recirculating aquaculture system located at northern China. Environ. Pollut. 2020, 261, 114155. [Google Scholar] [CrossRef]
- Lu, Z.; Na, G.; Gao, H.; Wang, L.; Bao, C.; Yao, Z. Fate of sulfonamide resistance genes in estuary environment and effect of anthropogenic activities. Sci. Total Environ. 2015, 527–528, 429–438. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Mende, D.R.; Waller, A.S.; Sunagawa, S.; Järvelin, A.I.; Chan, M.M.; Arumugam, M.; Raes, J.; Bork, P. Assessment of Metagenomic Assembly Using Simulated Next Generation Sequencing Data. PLoS ONE 2012, 7, e31386. [Google Scholar] [CrossRef]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E.; et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Ocean Plankton. Structure and function of the global ocean microbiome. Science 2015, 348, 794. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Villar, E.; Farrant, G.K.; Follows, M.; Garczarek, L.; Speich, S.; Audic, S.; Bittner, L.; Blanke, B.; Brum, J.R.; Brunet, C.; et al. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science 2015, 348, 1261447. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Huson, Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.-Q.; Li, B.; Ma, L.; Bao, P.; Zhou, X.; Zhang, T.; Zhu, Y.-G. Metagenomic profiles of antibiotic resistance genes in paddy soils from South China. FEMS Microbiol. Ecol. 2016, 92, fiw023. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring Variation of Antibiotic Resistance Genes in Activated Sludge over a Four-Year Period through a Metagenomic Approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Graat, E.A.; Haenen, A.P.; van Santen, M.G.; van Essen-Zandbergen, A.; Mevius, D.J.; van Duijkeren, E.; van Hoek, A.H. Extended-spectrum and AmpC β-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: Prevalence, risk factors and molecular characteristics. J. Antimicrob. Chemother. 2014, 69, 2669–2675. [Google Scholar] [CrossRef]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K.; et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 2011, 17, 1216–1222. [Google Scholar] [CrossRef]
- Voets, G.M.; Fluit, A.C.; Scharringa, J.; Schapendonk, C.; Munckhof, T.V.D.; Hall, M.A.L.-V.; Stuart, J.C. Identical plasmid AmpC beta-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int. J. Food Microbiol. 2013, 167, 359–362. [Google Scholar] [CrossRef]
- Miranda, C.D.; Tello, A.; Keen, P.L. Mechanisms of antimicrobial resistance in finfish aquaculture environments. Front. Microbiol. 2013, 4, 233. [Google Scholar] [CrossRef] [PubMed]
- Van Bambeke, F.; Balzi, E.; Tulkens, P.M. Antibiotic efflux pumps. Biochem. Pharmacol. 2000, 60, 457–470. [Google Scholar] [CrossRef]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Futur. Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Mazhar, S.H.; Li, X.; Rashid, A.; Su, J.; Xu, J.; Brejnrod, A.D.; Su, J.-Q.; Wu, Y.; Zhu, Y.-G.; Zhou, S.G.; et al. Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 2021, 755, 142702. [Google Scholar] [CrossRef]
- Hazards, E.; Panel, o.B.; Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; de Cesare, A.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar]
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Li, Y.; Wang, M.; Zeng, Z. Antibiotics, Antibiotic Resistance Genes, and Bacterial Community Composition in Fresh Water Aquaculture Environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Liu, P.; Jia, S.; He, X.; Zhang, X.; Ye, L. Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 2017, 188, 455–464. [Google Scholar] [CrossRef]
- Lin, S.-M.; Zhou, X.-M.; Zhou, Y.-L.; Kuang, W.-M.; Chen, Y.-J.; Luo, L.; Dai, F.-Y. Intestinal morphology, immunity and microbiota response to dietary fibers in largemouth bass, Micropterus salmoide. Fish Shellfish Immunol. 2020, 103, 135–142. [Google Scholar] [CrossRef]
- Ruzauskas, M.; Armalytė, J.; Lastauskienė, E.; Šiugždinienė, R.; Klimienė, I.; Mockeliūnas, R.; Bartkienė, E. Microbial and Antimicrobial Resistance Profiles of Microbiota in Common Carps (Cyprinus carpio) from Aquacultured and Wild Fish Populations. Animals 2021, 11, 929. [Google Scholar] [CrossRef]
- Wang, S.-T.; Meng, X.-Z.; Dai, Y.-F.; Zhang, J.-H.; Shen, Y.; Xu, X.-Y.; Wang, R.-Q.; Li, J.-L. Characterization of the intestinal digesta and mucosal microbiome of the grass carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 37, 100789. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Jogler, C. The bacterial phylum Planctomycetes as novel source for bioactive small molecules. Biotechnol. Adv. 2021, 53, 107818. [Google Scholar] [CrossRef] [PubMed]
- Kuebutornye, F.K.A.; Abarike, E.D.; Lu, Y. A review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yang, R.; Wu, Q.; Ding, Y.; Wang, Z.; Zhang, J.; Lei, T.; Wu, S.; Zhang, F.; Zhang, W.; et al. First report of the optrA-carrying multidrug resistance genomic island in Campylobacter jejuni isolated from pigeon meat. Int. J. Food. Microbiol. 2021, 354, 109320. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Gao, J.; Dai, H.; Wang, Z.; Zhao, Y.; Cui, Y. Higher spreading risk of antibacterial biocide and heavy metal resistance genes than antibiotic resistance genes in aerobic granular sludge. Environ. Res. 2022, 212, 113356. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Yang, Q.; Wang, R.; Wang, R.; Wang, Q.; Xin, Y. Evolution of Antibiotic Resistance and the Relationship between the Antibiotic Resistance Genes and Microbial Compositions under Long-Term Exposure to Tetracycline and Sulfamethoxazole. Int. J. Environ. Res. Public Health 2019, 16, 4681. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xin, Z.; Zhang, Y.; Chen, J.; Yan, J.; Li, H.; Hu, H. Long-term manure application increased the levels of antibiotics and antibiotic resistance genes in a greenhouse soil. Appl. Soil Ecol. 2017, 121, 193–200. [Google Scholar] [CrossRef]
- Pan, Z.; Yang, S.; Zhao, L.; Li, X.; Weng, L.; Sun, Y.; Li, Y. Temporal and spatial variability of antibiotics in agricultural soils from Huang-Huai-Hai Plain, northern China. Chemosphere 2021, 272, 129803. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Y.; Li, X.; Zhang, Y.; Ye, J.; Huang, H.; Zhu, C. Temporal effects of repeated application of biogas slurry on soil antibiotic resistance genes and their potential bacterial hosts. Environ. Pollut. 2020, 258, 113652. [Google Scholar] [CrossRef]
- Rahman, M.M.; Shan, J.; Yang, P.; Shang, X.; Xia, Y.; Yan, X. Effects of long-term pig manure application on antibiotics, abundance of antibiotic resistance genes (ARGs), anammox and denitrification rates in paddy soils. Environ. Pollut. 2018, 240, 368–377. [Google Scholar] [CrossRef]
- Song, J.; Shen, Q.; Wang, L.; Qiu, G.; Shi, J.; Xu, J.; Brookes, P.C.; Liu, X. Effects of Cd, Cu, Zn and their combined action on microbial biomass and bacterial community structure. Environ. Pollut. 2018, 243, 510–518. [Google Scholar] [CrossRef]
- Wang, X.; Lan, B.; Fei, H.; Wang, S.; Zhu, G. Heavy metal could drive co-selection of antibiotic resistance in terrestrial subsurface soils. J. Hazard. Mater. 2021, 411, 124848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, M.; Zhong, X.; Liu, P.; Xie, X.; Wangxiao, J.; Sun, Y. Dissemination of resistance genes in duck/fish polyculture ponds in Guangdong Province: Correlations between Cu and Zn and antibiotic resistance genes. Environ. Sci. Pollut. Res. 2019, 26, 8182–8193. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Lv, Z.; Shen, Y.; Liu, D.; Fu, Y.; Zhou, L.; Liu, W.; Chen, K.; Ye, H.; Xia, X.; et al. Metagenomic insights into differences in environmental resistome profiles between integrated and monoculture aquaculture farms in China. Environ. Int. 2020, 144, 106005. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).