Abstract
Litter size plays an essential role in mammalian evolution and is one of the most important factors determining whether an organism is deemed to have a ‘slow’ or ‘fast’ life history strategy. Humans are distinct in being classified as having slow life history yet bearing singletons who have completed relatively less growth than other ape neonates. Previous work has proposed that the ancestral primate gave birth to singletons. However, primate litter size has not yet been contextualized within a broad phylogenetic assessment of mammalian life history. We performed a comprehensive investigation of primate litter size using life history data for 155 primate species, and litter size data for an additional 791 boreoeutherian mammals. Litter size and life history traits have strong phylogenetic signal in primates (Pagel’s lambda: 0.99, p < 0.001; Blomberg’s K: 0.6311. p < 0.001), and litter size is significantly negatively correlated with gestation length (p < 0.001). Our data support that the last common ancestors of both primates and Haplorhini gave birth to multiples (litter size 1.7 and 1.6, respectively). We also find that singleton-bearing pregnancies evolved convergently in multiple primate lineages, including tarsiers and other haplorhines. This study contributes significantly to our understanding of life history and litter size in mammals, and we emphasize the utility of a callitrichid model for investigating the evolution of human reproduction.
Keywords:
litter size; phylogenetics; primates; Euarchonta; life history; ancestral state; Callitrichidae 1. Introduction
Almost all primates give birth to single offspring [1,2]. However, some genera, including marmosets, tamarins, lemurs, lorises, and galagos, regularly give birth to twins and even triplets [1,3,4,5].
While humans almost always give birth to singletons, twin pregnancies do occur spontaneously at a global rate of around 1.1–1.5% [6]. With advances in assisted reproductive technologies, twinning has increased in some regions to a rate of about 3.0% over the last 50 years [7]. It has become imperative to understand the effects of twinning on pregnancies, mothers, and newborns.
Litter size is an important characteristic of life history that influences many aspects of reproduction, growth, and development [8,9]. Offspring number impacts how much investment each individual can receive from parents and other community members, particularly when there are finite energies and resources available [10]. Classic life history theory, r/K selection theory [11], posits that unstable, unpredictable environments favor organisms with high litter sizes and rapid growth (r-selected organisms), while stable environments select for organisms with smaller litter sizes and higher resource investment (K-selected organisms).
A strict application of the r/K continuum is no longer common in part because it does not apply neatly in all cases [12]. It has since been replaced by a less binary framework describing life history trade-offs. The litter size trade-off involves trading the advantages of producing many offspring for the capacity to provide more prenatal and postnatal resources to each individual offspring [10,13]. These trade-offs function alongside selection to place organisms along a slow (similar but not identical to K-selected) to fast (r-selected) life history continuum [14]. Life history traits classically associated with slow life histories include small numbers of offspring (small litters), large neonatal and adult body mass, increased length of adolescence and maturation periods, among others [8,10].
Slow life histories and singleton litters have arisen multiple times in mammals, including within Homo sapiens [1]. This pattern of high investment in a single offspring has been associated with increased brain size [15] and prolonged maturation [16] in the primate lineage. In contrast, the production of single offspring in Chiroptera and Dermoptera has been associated with aerial locomotor requirements [17,18]. Broadly, increased litter size has been negatively correlated with body size dimorphism in a sample of non-primate mammals [19] and positively correlated with reduced body size in a sample of primates [20]. These findings suggest that larger litters are generally produced by smaller-bodied, less sexually dimorphic mammals.
Humans are a prime example of a species with a life history made up of both prototypically ‘slow’ and ‘fast’ traits. Humans generally produce singletons with large brain masses at birth [1,15] yet have a reduced interbirth interval compared to other extant apes [21]. This allows for humans to produce more offspring during their reproductive lifespans, despite investing heavily in each offspring. A critical understanding of how reproduction has evolved will help illuminate the population expansions and technological advances that humans have undergone over the last 200,000 years.
Because the vast majority of primates give birth to singletons, it has been theorized that the primate last common ancestor (LCA) also gave birth to singletons, and that platyrrhines and strepsirrhines have convergently evolved increased litter size [2,22]. However, many other mammals have litter sizes greater than one, and primate litter size has not been well contextualized within the broader mammalian phylogeny. To better understand the evolution of primate litter size, we collected data on litter size, gestation length (days), adult and neonatal brain and body mass (grams), and maximum longevity (years) for N = 946 boreoeutherians, including n = 155 primate species. To our knowledge, this is the most comprehensive phylogenetic analysis of primate litter size to date. Using a series of phylogenetic analyses and ancestral state reconstructions, we tested the following hypotheses:
H1.
Primate litter size is significantly associated with other life history and body size traits.
H1—null.
Litter size is not correlated with other life history and body size traits.
H2.
The primate last common ancestor had a litter size greater than 1.
H2—null.
The primate last common ancestor gave birth to singletons.
2. Materials and Methods
We collected data on litter size for N = 946 species of extant eutherian mammals across 106 families, including n = 155 primate species representing 16 families across all major lineages. For the primate species (n = 155), we collected additional data on other life history and body size traits. The breakdown of species and the references for the data are presented in the Supplementary Materials (primate dataset summarized in Table 1; see Table S1 for more information). We included representatives of all extant families within primates. More than 90% of all extant families are represented by an average litter size value for at least one species for every clade in Boreoeutheria; this includes all extant families in Scandentia and Dermoptera (collectively called Euarchonta alongside Primates).

Table 1.
Primate families and genera in the life history dataset.
We collected data on litter size as well as other life history and size traits, including gestation period in days, maximum longevity in years, age at female and male maturity in years, birth and adult weights measured in grams, litters per year, age at weaning in days, interbirth interval in days, and neonatal and adult brain mass in grams. See the full dataset in the Supplementary Materials (Table S1). All data were size-corrected (log10) prior to analyses. See the Supplementary Materials for our dataset, including a full species list and descriptive data for each primate family (Table S2).
We used previously published formulas to calculate prenatal growth rate (PGR, [31]), brain–body ratio (BBR, [27]), percentage of brain growth accomplished before birth (PGN, [32]), percentage of brain growth accomplished post-birth (PGV, [32]), and encephalization quotient (EQ, [33]):
2.1. Analytical Methods—Statistical Analyses
All analyses were performed using RStudio [34], on base R version 4.2.2 [35]. We first generated a series of summary statistics using the describeBy function in psych [36].
We then read in a phylogenetic tree of the sample using the read.tree function in ape [37]. We selected a tip-dated tree containing 5911 species, which was then trimmed to species represented in our sample [38]. The taxonomy used in this study follows the taxonomy of the phylogenetic tree [38] and does not necessarily reflect our personal views on mammalian taxonomy (e.g., family Hominidae; [39]).
We computed multivariate relationships between litter size, adult body mass, gestation length, brain mass, maximum longevity, brain–body ratio, and EQ with principal component analysis (PCA) using the prcomp function in the stats package built-in to R [35]. We visualized PC1–PC3 in bivariate space using qplot [40] and then vectorized the impact of each life history character on the PCs with the fviz_pca_var function in the factoextra package [41]. We visualized correlations between select life history variables using the cor function in stats [35] and the ggcorrplot function of the package ggcorrplot [42].
We used phylogenetic generalized least Squares (PGLS) analyses to assess the statistical relationship between litter size and every other life history and body size trait within a phylogenetic framework. Additionally, we performed PGLS analyses between all other traits used in the principal component analysis. All PGLS analyses were calculated using the pgls function in caper [43].
We calculated phylogenetic signal (Pagel’s Lambda, λ, and Blomberg’s K, [44]) for each life history and body/brain size trait using the function phylosig in phytools [45]. Phylogenetic signal tests are used to quantify the tendency of related biological species to resemble each other more than any other species. They produce a value which signifies how closely a given trait adheres to this tendency within a given phylogeny, comparing the character states for the trait present within the phylogeny and analyzing how closely the trait variation corresponds to phylogenetic relatedness.
Pagel’s lambda produces values between zero and one. When an estimated lambda equals zero, the traits are presumed to have no phylogenetic signal, supporting that the trait evolved independently of phylogenetic relatedness or might be under strong natural selection. A lambda of one corresponds to a Brownian motion model of evolution, and any other value is intermediate [46].
K variance differs. Brownian motion still produces an expected value of one, but K values range from greater to less than one. A K value under one is interpreted as taxa resembling each other less than expected, while a K greater than one is interpreted as taxa being more similar than Brownian motion would predict [44,47].
2.2. Analytical Methods—Ancestral State Reconstruction
We calculated two predictions of litter size for ancestral primates using ancestral state reconstruction (ASR; [18,48]). One ASR was conducted with primate data (n = 155 species) and a euarchontan outgroup (n = 8 species). To better contextualize the evolution of primate litter size within the broader mammalian phylogeny, we also collected data for an additional n = 783 species of extant eutherian mammals, including the majority (over 90%) of extant families, representing every clade within Boreoeutheria (total including primates, N = 946).
We reconstructed ancestral states for litter size using contMap and quantified the estimated values at internal nodes using fastAnc in phytools [45]. Ancestral state reconstruction values with associated node labels are available in Table S3 in the Supplementary Materials.
3. Results
Litter size has significant phylogenetic signal and is significantly correlated with gestation length in primates. Primates vary broadly in life history and body size (Table 2). Within this sample of 155 primates, adult body mass ranges from 42.5 g (Brown mouse lemur, Microcebus rufus) to over 120 kg (Western gorilla, Gorilla gorilla), mean litter size is generally close to 1 but goes up to 2.5 (M. rufus), gestation lengths are as short as 61 days (Gray mouse lemur, Microcebus murinus) and as long as 280 days (Homo sapiens), and maximum lifespan ranges from 13 (Calabar potto, Arctocebus calabarensis) to over 120 years (H. sapiens).

Table 2.
Mean values for selected life history characters across primate families.
3.1. Phylogenetic Signal
All life history and body size traits sampled here, including litter size, have highly significant phylogenetic signal (Table 3). The traits with the strongest phylogenetic signal in primates are litter size, prenatal growth rate, adult mass, and brain mass of both adults and neonates. The traits that show evidence of changing more rapidly within primates include encephalization quotient, brain–body ratio, and maximum longevity. Gestation length has an intermediate phylogenetic signal compared to most assessed traits. All traits are phylogenetically conserved.

Table 3.
Results of the phylogenetic signal analyses.
3.2. Phylogenetic Generalized Least Squares (PGLS) Analyses
All trait pairs compared here are significantly correlated (Table 4, Figure 1a). We find significant positive relationships between adult body and brain mass, adult body mass and maximum longevity, adult body mass and prenatal growth rate, adult body mass and gestation length, gestation length and adult brain mass, adult brain mass and maximum longevity, adult brain mass and prenatal growth rate, encephalization quotient and brain mass, maximum longevity and prenatal growth rate, maximum longevity and encephalization quotient, encephalization quotient and prenatal growth rate, and between brain–body ratio and encephalization quotient.

Table 4.
Results of the PGLS analyses.
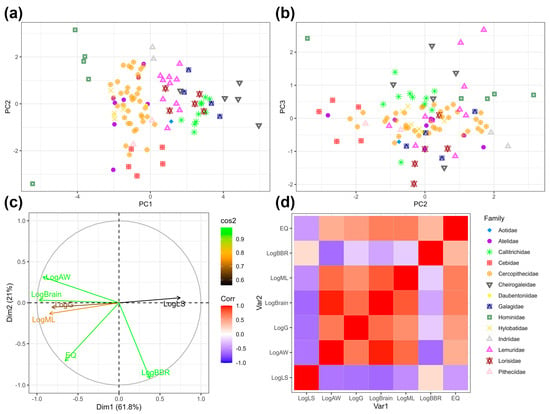
Figure 1.
Multivariate relationships between life history and body size traits in humans. Panels (a∓c) are based on a PCA including n = 101 primate species. Traits included in the PCA are litter size, adult weight, gestation length, brain mass, maximum longevity, brain–body ratio, and encephalization quotient. Primate families are designated by shapes and colors (see legend in figure). (a) PC1 v. PC2. (b) PC2 v. PC3. (c) Impact vectors for each life history variable. Dim1 = PC1, Dim2 = PC2. (d) Correlation plot of life history. Dark red colors indicate a strong positive correlation. Dark blue colors indicate a strong negative correlation.
Litter size and gestation length, adult body mass and brain–body ratio, adult body mass and encephalization quotient, and prenatal growth rate and brain–body ratio are all significantly negatively correlated in this sample.
The strongest statistical relationships in our sample are between adult brain and body mass, adult body mass and prenatal growth rate, adult brain mass and prenatal growth rate, adult body mass and brain–body ratio, and between brain–body ratio and encephalization quotient.
3.3. Principal Component Analyses (PCA) and Correlations
Body mass and life history traits clearly delineate primate families (Figure 1a,b). Hominids (all apes except the gibbons) sit in negative PC1 space, and the dwarf and mouse lemurs sit in positive PC1 space. Lemurs, lorises, and galagos sit in the same multivariate space. However, cebids sit outside the range of other platyrrhines.
PC2 reflects relative brain size and is highly impacted by brain–body ratio and encephalization (Figure 1c). Homo sapiens are on one end of the spectrum, with capuchins, squirrel monkeys, and spider monkeys closest to this extreme negative value of PC2. The four taxa closest to H. sapiens in PC2 space (Cebus albifrons, Ateles paniscus, Cebus capucinus, Saimiri sciureus) are all under 4.5 kg. On the other end is G. gorilla and other large-bodied primates with relatively smaller brains.
PC1 describes 61.8% of variance in the dataset, PC2 describes 21% of variance in the dataset, and PC3 describes 8.4% of variance in the dataset.
The vector biplot reinforces the statistical relationships between trait pairs. Litter size is negatively correlated with adult weight, gestation length, and maximum longevity. Brain body ratios and EQ are not as significantly correlated with other life history and body size traits (Figure 1c,d).
3.4. Ancestral State Reconstruction—Boreoeutherian Dataset
We reconstructed ancestral primate litter size using two different datasets. Using only euarchontan taxa, the ancestral primate is supported to have given birth to singletons (Figure 2). However, when we contextualize primate litter size with the full boreoeutherian dataset (n = 955), the ancestral primate is supported to have given birth to twins (litter size 1.7, Table 5, Figure 3). This litter size value is closest to the extant tamarin species Saguinus bicolor, the pied tamarin (Callitrichidae).
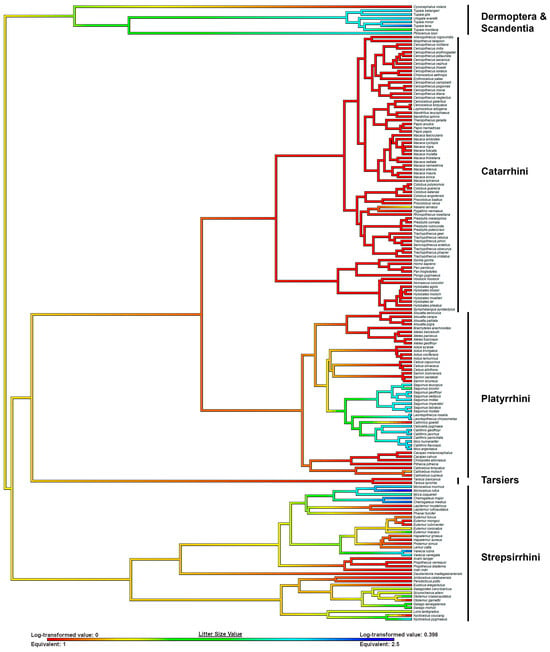
Figure 2.
Ancestral state reconstruction (ASR) of litter size in Euarchonta. Litter size values are beneath the ASR. Warmer colors indicate smaller litter sizes, and cooler colors indicate larger litter sizes. Major clades are labeled.

Table 5.
Various ancestral state reconstructions of litter size.
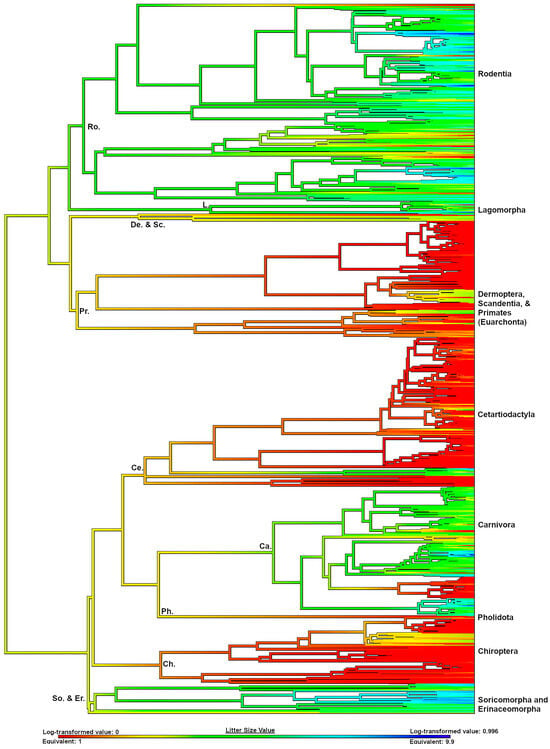
Figure 3.
Ancestral state reconstruction (ASR) of litter size in Boreoeutheria. Litter size values are beneath the ASR. Warmer colors indicate smaller litter sizes, and cooler colors indicate larger litter sizes. Major clades are labeled.
Using a comprehensive mammalian phylogeny, litter size data support that strepsirrhines evolved from a twinning ancestor, and twinning in that lineage is an ancestral trait. Tarsiers are also supported to have evolved from a twinning ancestor, a finding that is bolstered by the presence of supernumerary nipples in extant species. Our reconstruction supports that singleton births evolved after the divergence and diversification of haplorhines approximately 45–65 million years ago [49]. Twinning platyrrhines are supported to have re-evolved a multifetal reproductive strategy in the Miocene (Table 6, see more information in Table S3).

Table 6.
Litter size values for major primate lineage last common ancestors (LCAs).
4. Discussion
A phylogenetically comprehensive investigation of litter size supports that the earliest primates likely gave birth to twins. Primates are characterized by relatively large brains, as well as relatively long lifespans and other developmental periods (maturation times, gestation). Our findings highlight the unique life history strategy of humans relative to other nonhuman primates. Despite the tight connection between prenatal growth rate, adult body mass, and small litter size, humans have small litter sizes alongside a relatively small percentage of brain growth accomplished prenatally (less than 25%, secondarily altricial). This contrasts with other primates like Western tarsiers (Tarsius bancanus) and Western gorillas (G. gorilla), which give birth to singletons that accomplish over 60% of brain growth before birth [1]. Humans also demonstrate a brain–body ratio and relative lifespan similar to much smaller-bodied species like capuchins.
Life history traits within primates covary along lines predicted by theories of a slow–fast life history continuum [14]. Life history and body size are highly integrated in primates and likely coevolved, leading to a continuum where organisms tend to have slower or faster life history ‘versions’ of many traits. These trends in life history trait covariation across Primates support those previously identified in other studies [20] and speak to the existence of extrinsic life history constraints which can be used to inform our understanding of evolutionary change. Primates are well known to have smaller litter sizes than other eutherian orders [12], so it is not surprising that the primate ancestor is supported to have gestated multifetal litters. The evolutionary reduction in primate litter size may be related to changes in environment or social structure that enabled greater investment in fewer offspring (slow life history; [14]).
4.1. Evolution of Primate Litter Size
The primate clade is estimated to have originated 60–70 mya [38,49,51] around the Cretaceous–Paleogene (K-Pg) mass extinction event. The earliest plesiadapiform fossils are from present-day North America [52], and the earliest primate fossil was discovered in present-day China [53]. Previous reconstructions of ancestral litter size have focused on primate and euarchontan lineages. Our broader taxonomic assessment of litter size supports that the ancestral primate gave birth to twins.
Because litter size has a strong phylogenetic signal, assessing this life history trait across the mammalian phylogeny gives a better picture of the ancestral state and can shed light on the evolutionary processes shaping human evolution. Novel to this work, an average litter size of approximately two is predicted for the last common ancestor of primates dated between approximately 77 and 60 million years ago [38,49,51]. The singletons most common in extant members of our order appear to have not been selectively advantageous until more recently in our evolutionary history. Early primate fossil anatomy (e.g., Archicebus achilles) aligns with our work and indicates that early primates were similar in body size to small, twinning primates like Microcebus [53]. The presence of three nipple pairs in both some strepsirrhine and some haplorhine taxa [48] also supports a larger ancestral litter size.
Nipple number and litter size are tightly correlated in primates [54]. Several primates, including some tarsiers and strepsirrhines (e.g., Philippine tarsiers, Tarsius syrichta, and Calabar pottos, A. calabarensis; [48]), have more than two pairs of nipples, a trait typically associated with larger litters. Our results support previous studies arguing that twinning strepsirrhines retain multifetal litters from a common ancestor. Our results also support that all primates share an ancestor that gave birth to multiples. The evolution of singletons in primates may have evolved separately in simians (monkeys and apes) and tarsiers. We reconstruct the LCA of haplorhines to have a litter size of 1.56 (node 1488). This convergent evolution of singletons could shed light on why tarsiers retain multiple pairs of nipples.
Placentation may also be linked to litter size and gestation length. Invasive placentation was the likely ancestral condition of both Primates and Euarchontoglires, and epitheliochorial placentation likely coevolved with twinning in extant strepsirrhines [21]. Future work investigating the phenotypic relationship between placental type, nipple number, litter sizes, and gestation length may shed additional light on the interactions between these reproductive traits.
During the evolution of Boreoeutheria, lineages experienced two mass extinction events, one at the Cretaceous–Paleogene boundary and the other at the Eocene–Oligocene boundary [55]. Twinning appears to have been the likely condition for the last common ancestor of primates which lived prior to at least the second extinction event and possibly even prior to the first extinction event which occurred around 66 million years ago [56]. Giving birth to multiples may have been an important factor defining the life history strategy which allowed our ancestors and those of other extant primate lineages to survive these extinction events. Additionally, these massive ecological shifts may have opened novel niches for primates with slower life histories, which allowed for singletons to evolve as an adaptive strategy during a period of climatic stability.
4.2. Callitrichids as a Model for the Evolution of Human Reproduction
Callitrichids are commonly used as a model for understanding human evolution, (e.g., as models for the evolution of phyletic nanism in Homo floresiensis [31]), as they are social primates that score very high on cognitive tests [57]. Additionally, callitrichids practice pair-bonding and alloparental care, with parents and other adults offering care for the young [3,5,57,58,59]. Callitrichids have been reported as the only group which practices cooperative breeding to an extent comparable with humans and, combined with twinning, have a higher theoretical maximum reproductive rate than other primates [60]. In addition to the prevalence of twinning in this family, some callitrichid populations regularly birth two litters per year [61], and triplets are the most prevalent litter size in some populations of common marmosets (Callithrix jacchus) [62].
Like humans, callitrichids are frequently classified as ‘secondarily altricial’, as their neonates have completed proportionally less of their brain and body growth than other closely related monkeys [59]. This results directly from their slow fetal growth rate [63], likely associated with regular twinning in many taxa in this family [5,58]. In humans, twinning is also associated with slow fetal growth rates, particularly in the third trimester [64]. As fetal growth rates impact craniofacial development, including tooth proportions and adult brain size [63,65], understanding the mechanisms surrounding multifetal pregnancies has significant implications for modern human health as well as human evolution.
Our work reinforces the utility of a callitrichid model for investigating the evolution of human reproduction and life history. Evolutionary increases in hominid brain size were likely tightly associated with pair-bonding and alloparental care [59,60,65,66]. Alloparental care, whether this is derived from pair-bonding or an alternative social system [67], may be key to supporting multiple fetuses and other costly pregnancies in primates. The success of the human species, which is often measured in brain/body size ratios and cognitive abilities, seems inextricable from changes in reproductive strategy and energy allocation throughout evolutionary history [65,68]. The earliest primates likely gave birth to twins, and twinning in modern humans may bear genetic and developmental links with those ancestors. We highlight the growing evidence for tight links between reproduction and brain/body size traits, and we argue for continued investigation of these correlations in callitrichids and other mammalian species.
Our reconstructions support that the primate and haplorhine last common ancestors may have been twinning species, and investing in singletons may have become the norm more recently. Our work supports recent reconstructions arguing that primate social organization began with flexible pair-bonding rather than solitary living [69]. Likewise, our reconstruction of twinning may suggest that alloparental (or at least allomaternal) care and relatively cohesive social groupings defined the earliest primates. The negative relationship between litter size and gestation length specifically speaks to the constraints imposed upon reproductive variation by the prenatal requirements of offspring, which are less dispersible than postnatal requirements. Our finding that litter size is not significantly correlated with other life history traits within primates may be related to the derived ability of primates to disperse postnatal care requirements through shared childcare burdens.
Human reproductive output far outpaces our great ape relatives due to our reduced interbirth intervals, made possible in part by alloparental care. Callitrichids not only offer a potential comparison to the earliest primates but similarly show relatively higher reproductive output and alloparental care compared to their closest relatives (Cebidae, the squirrel, and capuchin monkeys, [59,60]). Further investigation into the role of twinning in human reproduction may benefit from looking to callitrichid monkeys as model organisms. Of special interest is whether apes do indeed twin more often than other catarrhine primates, as has been suggested [70], especially considering the possible fitness impacts of twinning (both positive and negative) in humans [71,72]. Convergent evolution of alloparental care, cooperative behavior patterns, and possibly twinning in callitrichids and humans may shed light on critical events in human evolution, including the Miocene ape radiation and the origins of genus Homo.
5. Conclusions
Litter size plays an important role in the life history strategy of all mammalian taxa. In humans, giving birth to twins occurs at a low but stable frequency that is increasing with more wide-spread use of assisted reproductive technologies. Our work demonstrates that litter size has a strong and significant phylogenetic signal in primates and is significantly correlated with gestation length. Multifetal primate pregnancies are significantly associated with small brain and body size, short gestation, and rapid growth. Humans, in contrast, have long lifespans, slow growth, long gestation, large brain and body size, and almost always give birth to singletons.
Reconstructing primate litter size within the context of other mammals supports twinning as the ancestral condition at the origin of the primate lineage around 60–70 million years ago. A reproductive strategy of giving birth to singletons likely coevolved with longer gestations, and larger brain and body size, in many primate lineages, including humans. Evolution of singletons likely occurred early in the primate lineage, but after the divergence of strepsirrhines approximately 55 mya, and singletons may have coevolved in simians and tarsiers. Further work incorporating life history data across mammals more broadly may help elucidate the evolutionary pressures driving changes in litter size. We also highlight the tight correlations between life history and body size traits, and we emphasize the benefits of a callitrichid model for investigating the evolution of human reproduction.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/humans4030014/s1; Table S1: Full dataset; Table S2: Descriptive data; Table S3: Node values for ASR; Table S4: Full phylogenetic signal results; Table S5: Full phylogenetic generalized least squares (PGLS) results.
Author Contributions
Conceptualization, J.H.M. and T.A.M.; methodology, J.H.M. and T.A.M.; software, J.H.M. and T.A.M.; validation, J.H.M. and T.A.M.; formal analysis, J.H.M. and T.A.M.; investigation, J.H.M. and T.A.M.; resources, J.H.M. and T.A.M.; data curation, J.H.M.; writing—original draft preparation, J.H.M.; writing—review and editing, J.H.M. and T.A.M.; visualization, J.H.M. and T.A.M.; supervision, T.A.M.; project administration, T.A.M.; funding acquisition, J.H.M. and T.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Western Washington University (GGR05F and GGR06C to J.H.M) and the Leakey Foundation (to T.A.M).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article and Supplementary Materials; further inquiries can be directed to the corresponding author.
Acknowledgments
We would like to thank the researchers who made these life history and body size data available. In addition, we would like to thank A. Weitz, H. Schwandt, and T. Koetje for their support and mentorship during the research process.
Conflicts of Interest
J.H.M. has received funding from Western Washington University; T.A.M. has received funding from the Leakey Foundation. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results. All authors declare no conflicts of interest.
References
- Ernest, S.K.M. Life History Characteristics of Placental Nonvolant Mammals: Ecological Archives E084-093. Ecology 2003, 84, 3402. [Google Scholar] [CrossRef]
- Leutenegger, W. Evolution of Litter Size in Primates. Am. Nat. 1979, 114, 525–531. [Google Scholar] [CrossRef]
- Dunbar, R.I.M. The Mating System of Callitrichid Primates: I. Conditions for the Coevolution of Pair Bonding and Twinning. Anim. Behav. 1995, 50, 1057–1070. [Google Scholar] [CrossRef]
- Martin, R.D. Reproductive Characteristics of New World Monkeys. Int. Zoo Yearb. 2012, 46, 95–108. [Google Scholar] [CrossRef]
- Ross, C.N.; Fite, J.E.; Jensen, H.; French, J.A. Demographic Review of a Captive Colony of Callitrichids (Callithrix kuhlii). Am. J. Primatol. 2007, 69, 234–240. [Google Scholar] [CrossRef][Green Version]
- Blickstein, I.; Keith, L.G. Multiple Pregnancy: Epidemiology, Gestation & Perinatal Outcome, 2nd ed.; Taylor & Francis: London, UK, 2005; ISBN 978-1-84214-239-4. [Google Scholar]
- Ananth, C.V.; Chauhan, S.P. Epidemiology of Twinning in Developed Countries. Semin. Perinatol. 2012, 36, 156–161. [Google Scholar] [CrossRef]
- Frynta, D.; Fraňková, M.; Čížková, B.; Skarlandtová, H.; Galeštoková, K.; Průšová, K.; Šmilauer, P.; Šumbera, R. Social and Life History Correlates of Litter Size in Captive Colonies of Precocial Spiny Mice (Acomys). Acta Theriol. (Warsz.) 2011, 56, 289–295. [Google Scholar] [CrossRef]
- Itô, Y.; Iwasa, Y. Evolution of Litter Size. Res. Popul. Ecol. 1981, 23, 344–359. [Google Scholar] [CrossRef]
- Walker, R.S.; Gurven, M.; Burger, O.; Hamilton, M.J. The Trade-Off between Number and Size of Offspring in Humans and Other Primates. Proc. Biol. Sci. 2008, 275, 827–833. [Google Scholar] [CrossRef]
- MacArthur, R.H. The Theory of Island Biogeography; Monographs in Population Biology; Princeton University Press: Princeton, NJ, USA, 1967. [Google Scholar]
- Stearns, S.C.; Koella, J.C. The Evolution of Phenotypic Plasticity in Life-History Traits: Predictions of Reaction Norms for Age and Size at Maturity. Evolution 1986, 40, 893–913. [Google Scholar] [CrossRef]
- Pagel, M.D.; Harvey, P.H. How Mammals Produce Large-Brained Offspring. Evolution 1988, 42, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992; ISBN 9780198577416. [Google Scholar]
- DeSilva, J.M.; Lesnik, J.J. Brain Size at Birth throughout Human Evolution: A New Method for Estimating Neonatal Brain Size in Hominins. J. Hum. Evol. 2008, 55, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Aiello, L.C.; Wells, J.C.K. Energetics and the Evolution of the Genus Homo. Annu. Rev. Anthropol. 2002, 31, 323–338. [Google Scholar] [CrossRef]
- Fokidis, H.B.; Risch, T.S. The Burden of Motherhood: Gliding Locomotion in Mammals Influences Maternal Reproductive Investment. J. Mammal. 2008, 89, 617–625. [Google Scholar] [CrossRef]
- Garbino, G.S.T.; Feijó, A.; Beltrão-Mendes, R.; Da Rocha, P.A. Evolution of Litter Size in Bats and Its Influence on Longevity and Roosting Ecology. Biol. J. Linn. Soc. 2021, 132, 676–684. [Google Scholar] [CrossRef]
- Carranza, J. Sexual Selection for Male Body Mass and the Evolution of Litter Size in Mammals. Am. Nat. 1996, 148, 81–100. [Google Scholar] [CrossRef]
- Harvey, P.H.; Clutton-Brock, T.H. Life History Variation in Primates. Evolution 1985, 39, 559–581. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.E. Comparative Reproductive Energetics of Human and Nonhuman Primates. Annu. Rev. Anthropol. 2013, 42, 287–304. [Google Scholar] [CrossRef]
- Leutenegger, W. Maternal-Fetal Weight Relationships in Primates. Folia Primatol. 1973, 20, 280–293. [Google Scholar] [CrossRef]
- Burger, J.R.; George, M.A., Jr.; Leadbetter, C.; Shaikh, F. The Allometry of Brain Size in Mammals. J. Mammal. 2019, 100, 276–283. [Google Scholar] [CrossRef]
- Capellini, I.; Venditti, C.; Barton, R.A. Placentation and Maternal Investment in Mammals. Am. Nat. 2011, 177, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Fredrickson, D. Hexaprotodon liberiensis (Madagascan Pygmy Hippopotamus). Available online: https://animaldiversity.org/accounts/Hexaprotodon_liberiensis/ (accessed on 1 February 2023).
- Ridgway, S.H.; Carlin, K.P.; Van Alstyne, K.R. Delphinid Brain Development from Neonate to Adulthood with Comparisons to Other Cetaceans and Artiodactyls. Mar. Mammal Sci. 2018, 34, 420–439. [Google Scholar] [CrossRef]
- Sacher, G.A.; Staffeldt, E.F. Relation of Gestation Time to Brain Weight for Placental Mammals: Implications for the Theory of Vertebrate Growth. Am. Nat. 1974, 108, 593–615. [Google Scholar] [CrossRef]
- Shefferly, N. Papio papio (Guinea Baboon). Available online: https://animaldiversity.org/accounts/Papio_papio/ (accessed on 1 February 2023).
- Shefferly, N. Papio anubis (Anubis Baboon). Available online: https://animaldiversity.org/accounts/Papio_anubis/ (accessed on 1 February 2023).
- Tacutu, R.; Thornton, D.; Johnson, E.; Budovsky, A.; Barardo, D.; Craig, T.; Diana, E.; Lehmann, G.; Toren, D.; Wang, J.; et al. Human Ageing Genomic Resources: New and Updated Databases. Nucleic Acids Res. 2018, 46, D1083–D1090. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.H.; Mundy, N.I. Parallel Episodes of Phyletic Dwarfism in Callitrichid and Cheirogaleid Primates. J. Evol. Biol. 2013, 26, 810–819. [Google Scholar] [CrossRef] [PubMed]
- Eisert, R.; Potter, C.W.; Oftedal, O.T. Brain Size in Neonatal and Adult Weddell Seals: Costs and Consequences of Having a Large Brain. Mar. Mammal Sci. 2014, 30, 184–205. [Google Scholar] [CrossRef]
- Jerison, H.J. Animal Intelligence as Encephalization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1985, 308, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Posit Team RStudio: Integrated Development Environment for R. 2022. Available online: http://www.posit.co/ (accessed on 25 February 2023).
- R Core Team. R: A Language and Environment for Statistical Computing. 2022. Available online: https://www.R-project.org/ (accessed on 25 February 2023).
- Revelle, W. Psych: Procedures for Psychological, Psychometric, and Personality Research. 2023. Available online: https://CRAN.R-project.org/package=psych (accessed on 23 March 2023).
- Paradis, E.; Blomberg, S.; Bolker, B.; Brown, J.; Claramunt, S.; Claude, J.; Cuong, H.S.; Desper, R.; Didier, G.; Durand, B.; et al. Ape: Analyses of Phylogenetics and Evolution. 2022. Available online: https://CRAN.R-project.org/package=ape (accessed on 2 February 2023).
- Upham, N.S.; Esselstyn, J.A.; Jetz, W. Inferring the Mammal Tree: Species-Level Sets of Phylogenies for Questions in Ecology, Evolution, and Conservation. PLoS Biol. 2019, 17, e3000494. [Google Scholar] [CrossRef]
- Johanson, D.C.; White, T.D. A Systematic Assessment of Early African Hominids. Science 1979, 203, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://cloud.r-project.org/web/packages/factoextra/index.html (accessed on 14 March 2024).
- Kassambara, A.; Patil, I. Ggcorrplot: Visualization of a Correlation Matrix Using “Ggplot2”. 2023. Available online: https://cran.r-project.org/web/packages/ggcorrplot/index.html (accessed on 14 March 2024).
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Pearse, W. Caper: Comparative Analyses of Phylogenetics and Evolution in R. 2018. Available online: https://CRAN.R-project.org/package=caper (accessed on 2 February 2023).
- Diniz-Filho, J.A.F.; Santos, T.; Rangel, T.F.; Bini, L.M. A Comparison of Metrics for Estimating Phylogenetic Signal under Alternative Evolutionary Models. Genet. Mol. Biol. 2012, 35, 673–679. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things): Phytools: R Package. 2012. Available online: https://onlinelibrary.wiley.com/doi/10.1111/j.2041-210X.2011.00169.x (accessed on 2 February 2023).
- Molina-Venegas, R.; Rodríguez, M.Á. Revisiting Phylogenetic Signal; Strong or Negligible Impacts of Polytomies and Branch Length Information? BMC Evol. Biol. 2017, 17, 53. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, S.P.; Garland, T., Jr.; Ives, A.R. Testing for Phylogenetic Signal in Comparative Data: Behavioral Traits Are More Labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef] [PubMed]
- Kappeler, P.M. Nests, Tree Holes, and the Evolution of Primate Life Histories. Am. J. Primatol. 1998, 46, 7–33. [Google Scholar] [CrossRef]
- Pozzi, L.; Hodgson, J.A.; Burrell, A.S.; Sterner, K.N.; Raaum, R.L.; Disotell, T.R. Primate Phylogenetic Relationships and Divergence Dates Inferred from Complete Mitochondrial Genomes. Mol. Phylogenet. Evol. 2014, 75, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.C.; Lynch Alfaro, J.W.; Rylands, A.B.; Alfaro, M.E. Biogeography of the Marmosets and Tamarins (Callitrichidae). Mol. Phylogenet. Evol. 2015, 82, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Steiper, M.E.; Young, N.M. Primate Molecular Divergence Dates. Mol. Phylogenet. Evol. 2006, 41, 384–394. [Google Scholar] [CrossRef]
- Wilson Mantilla, G.P.; Chester, S.G.B.; Clemens, W.A.; Moore, J.R.; Sprain, C.J.; Hovatter, B.T.; Mitchell, W.S.; Mans, W.W.; Mundil, R.; Renne, P.R. Earliest Palaeocene Purgatoriids and the Initial Radiation of Stem Primates. R. Soc. Open Sci. 2021, 8, 210050. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Gebo, D.L.; Dagosto, M.; Meng, J.; Tafforeau, P.; Flynn, J.J.; Beard, K.C. The Oldest Known Primate Skeleton and Early Haplorhine Evolution. Nature 2013, 498, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Rawal, D. Relationship between Number of Teats and Litter Size in Eutherian Mammals. Int. J. Sci. Res. Biol. Sci. 2019, 6, 249–252. [Google Scholar] [CrossRef]
- Wu, J.; Yonezawa, T.; Kishino, H. Evolution of Reproductive Life History in Mammals and the Associated Change of Functional Constraints. Genes 2021, 12, 740. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, S.; Xu, J.; Chen, B.; Zhou, K.; Yang, G. Phylogenomic Analysis Resolves the Interordinal Relationships and Rapid Diversification of the Laurasiatherian Mammals. Syst. Biol. 2012, 61, 150. [Google Scholar] [CrossRef] [PubMed]
- Burkart, J.M.; van Schaik, C.P. Cognitive Consequences of Cooperative Breeding in Primates? Anim. Cogn. 2010, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- McCoy, D.E.; Frye, B.M.; Kotler, J.; Burkart, J.M.; Burns, M.; Embury, A.; Eyre, S.; Galbusera, P.; Hooper, J.; Idoe, A.; et al. A Comparative Study of Litter Size and Sex Composition in a Large Dataset of Callitrichine Monkeys. Am. J. Primatol. 2019, 81, e23038. [Google Scholar] [CrossRef] [PubMed]
- Isler, K.; van Schaik, C.P. Allomaternal Care, Life History and Brain Size Evolution in Mammals. J. Hum. Evol. 2012, 63, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Isler, K.; van Schaik, C.P. How Our Ancestors Broke through the Gray Ceiling: Comparative Evidence for Cooperative Breeding in Early Homo. Curr. Anthropol. 2012, 53, S453–S465. [Google Scholar] [CrossRef]
- Godoy, I.; Perry, S.E. Mating Systems of New World Monkeys. In Encyclopedia of Animal Behavior, 2nd ed.; Choe, J.C., Ed.; Academic Press: Oxford, UK, 2019; pp. 563–567. ISBN 978-0-12-813252-4. [Google Scholar]
- Rutherford, J.N.; Ross, C.N.; Ziegler, T.; Burke, L.A.; Steffen, A.D.; Sills, A.; Layne Colon, D.; deMartelly, V.A.; Narapareddy, L.R.; Tardif, S.D. Womb to Womb: Maternal Litter Size and Birth Weight but Not Adult Characteristics Predict Early Neonatal Death of Offspring in the Common Marmoset Monkey. PLoS ONE 2021, 16, e0252093. [Google Scholar] [CrossRef] [PubMed]
- Monson, T.A.; Coleman, J.L.; Hlusko, L.J. Craniodental Allometry, Prenatal Growth Rates, and the Evolutionary Loss of the Third Molars in New World Monkeys. Anat. Rec. 2019, 302, 1419–1433. [Google Scholar] [CrossRef] [PubMed]
- Stirrup, O.T.; Khalil, A.; D’Antonio, F.; Thilaganathan, B.; Collaborative (STORK), on behalf of the S.T.O.R. Fetal Growth Reference Ranges in Twin Pregnancy: Analysis of the Southwest Thames Obstetric Research Collaborative (STORK) Multiple Pregnancy Cohort. Ultrasound Obstet. Gynecol. 2015, 45, 301–307. [Google Scholar] [CrossRef]
- Heldstab, S.A.; Isler, K.; Burkart, J.M.; van Schaik, C.P. Allomaternal Care, Brains and Fertility in Mammals: Who Cares Matters. Behav. Ecol. Sociobiol. 2019, 73, 71. [Google Scholar] [CrossRef]
- Monson, T.A.; Weitz, A.P.; Brasil, M.B.; Hlusko, L.J. Teeth, Prenatal Growth Rates, and the Evolution of Human-Like Pregnancy in Later Homo. Proc. Nat. Acad. Sci. USA 2022, 119, e2200689119. [Google Scholar] [CrossRef]
- Savage, A.; Snowdon, C.T.; Soto, L.; Medina, F.; Emeris, G.; Guillen, R. Factors Influencing the Survival of Wild Cotton-Top Tamarin (Saguinus oedipus) Infants. Am. J. Primatol. 2021, 83, e23262. [Google Scholar] [CrossRef] [PubMed]
- Brasil, M.F.; Monson, T.A.; Schmitt, C.A.; Hlusko, L.J. A Genotype: Phenotype Approach to Testing Taxonomic Hypotheses in Hominids. Sci. Nat. 2020, 107, 40. [Google Scholar] [CrossRef] [PubMed]
- Olivier, C.-A.; Martin, J.S.; Pilisi, C.; Agnani, P.; Kauffmann, C.; Hayes, L.; Jaeggi, A.V.; Schradin, C. Primate Social Organization Evolved from a Flexible Pair-Living Ancestor. Proc. Natl. Acad. Sci. USA 2024, 121, e2215401120. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, T. Twinning Frequency in Catarrhine Primates. Hum. Evol. 1990, 5, 387–396. [Google Scholar] [CrossRef]
- Gabler, S.; Voland, E. Fitness of Twinning. Hum. Biol. 1994, 66, 699–713. [Google Scholar] [PubMed]
- Lummaa, V.; Jokela, J.; Haukioja, E. Gender Difference in Benefits of Twinning in Pre-Industrial Humans: Boys Did Not Pay. J. Anim. Ecol. 2001, 70, 739–746. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
