Radiomic-Based Machine Learning for Differentiating Brain Metastases Recurrence from Radiation Necrosis Post-Gamma Knife Radiosurgery: A Feasibility Study
Abstract
1. Introduction
1.1. Radiomics and Machine Learning
1.2. Support Vector Machine
2. Materials and Methods
2.1. Software and Tools Used
2.2. Data Sources and Preparation
2.3. Radiomic Feature Extraction and Analysis
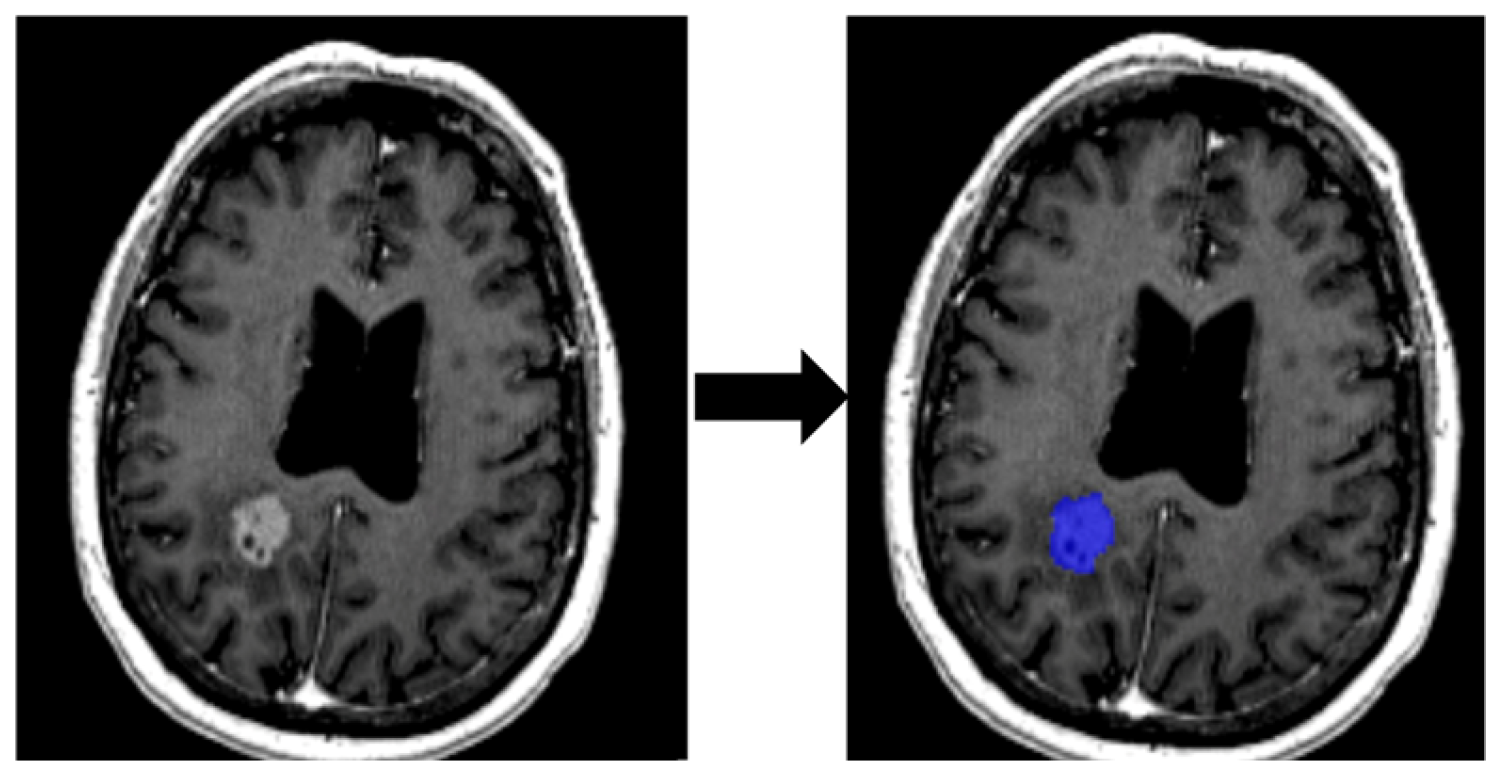
2.3.1. Segmentation of ROI
2.3.2. Feature Extraction
2.3.3. Preprocessing and Training
2.3.4. Model Selection
3. Results
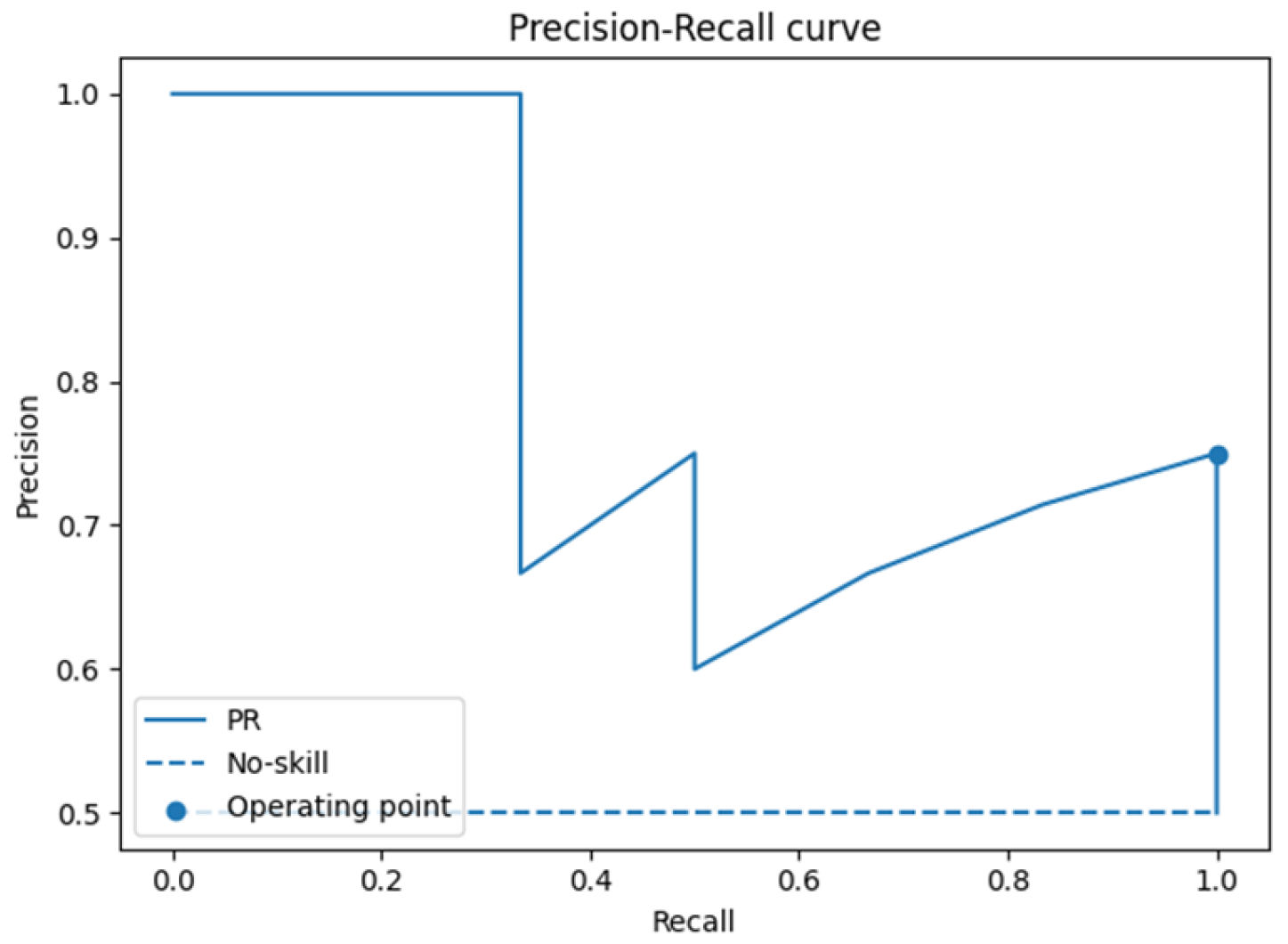
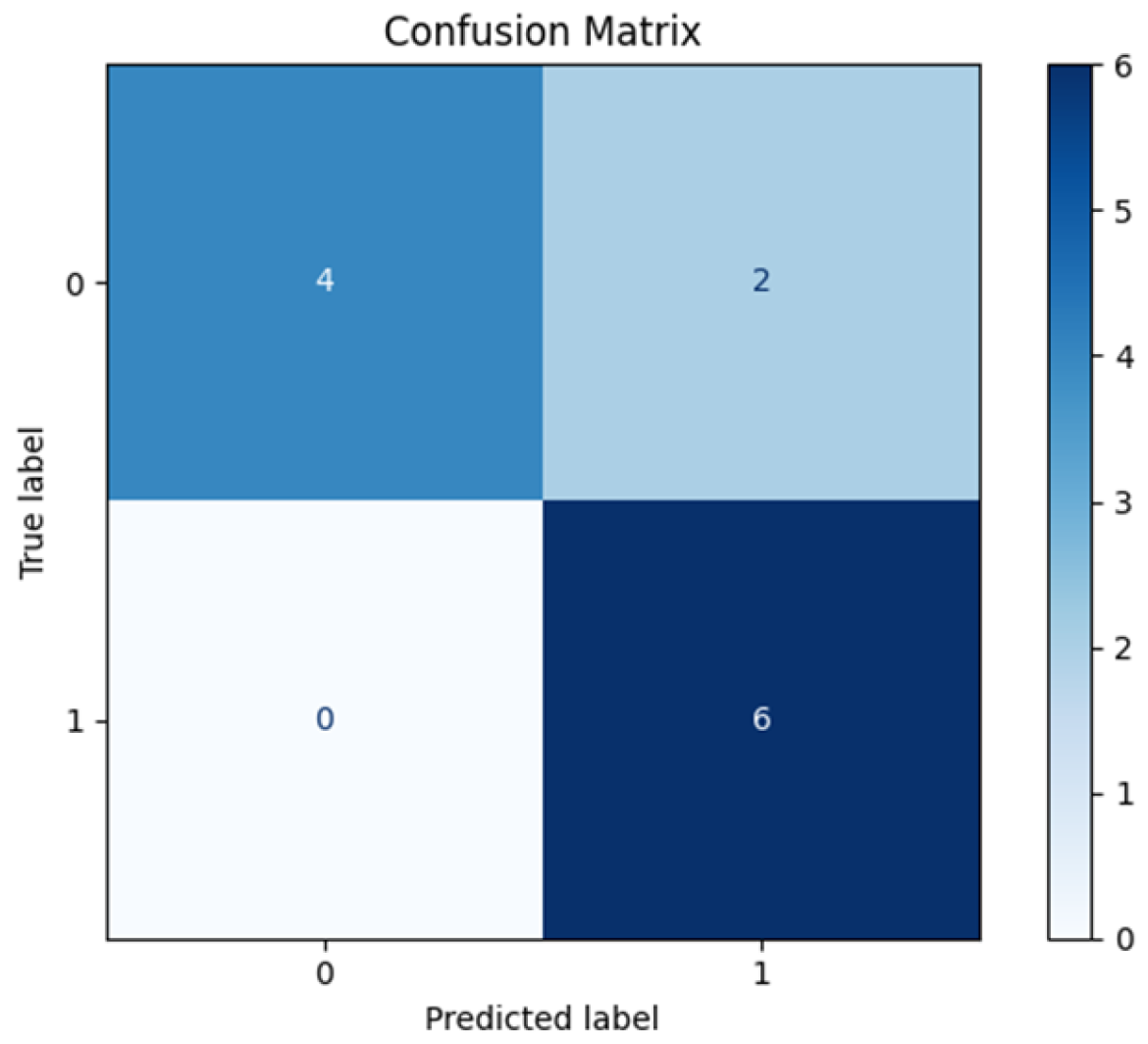
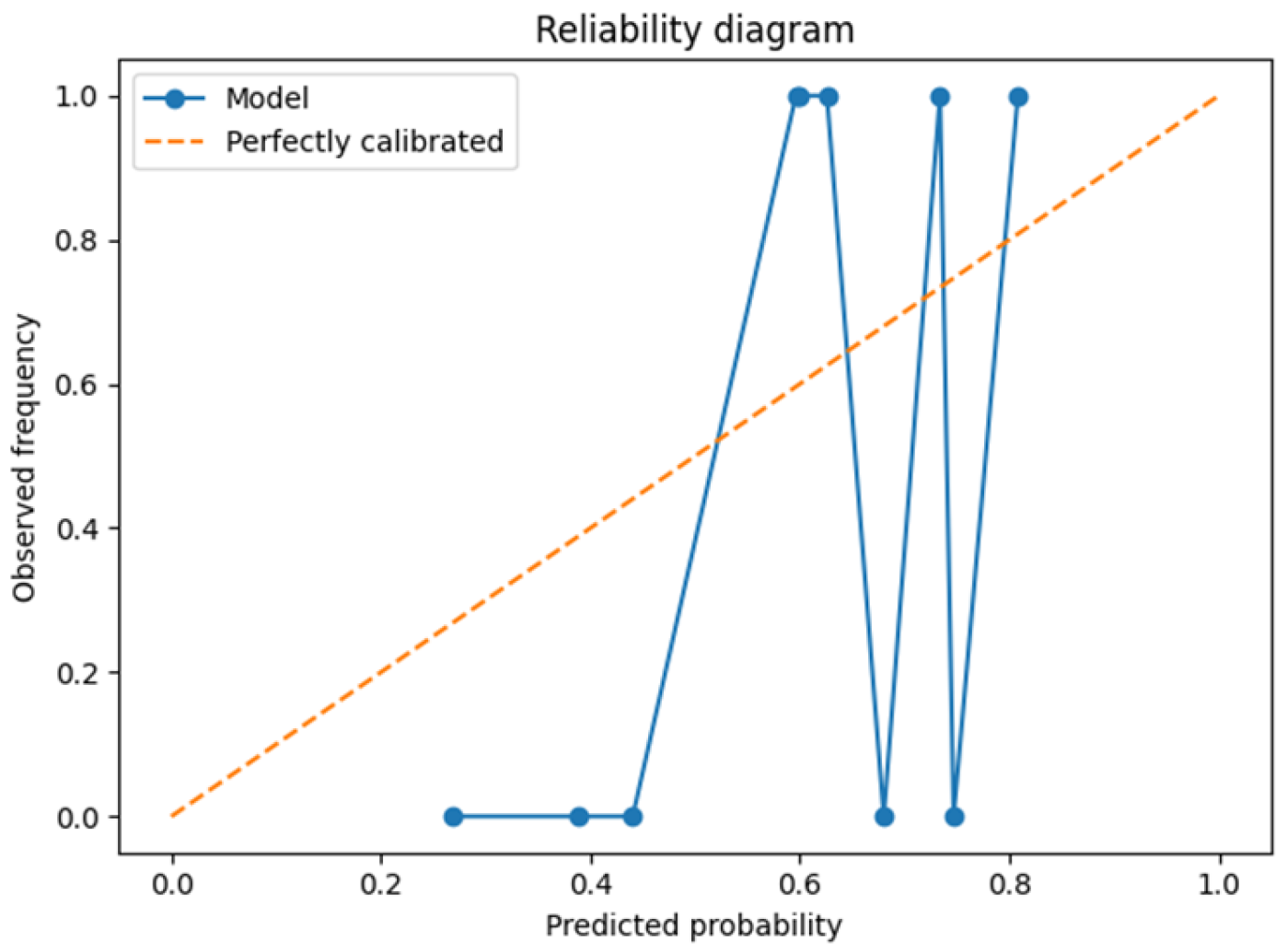
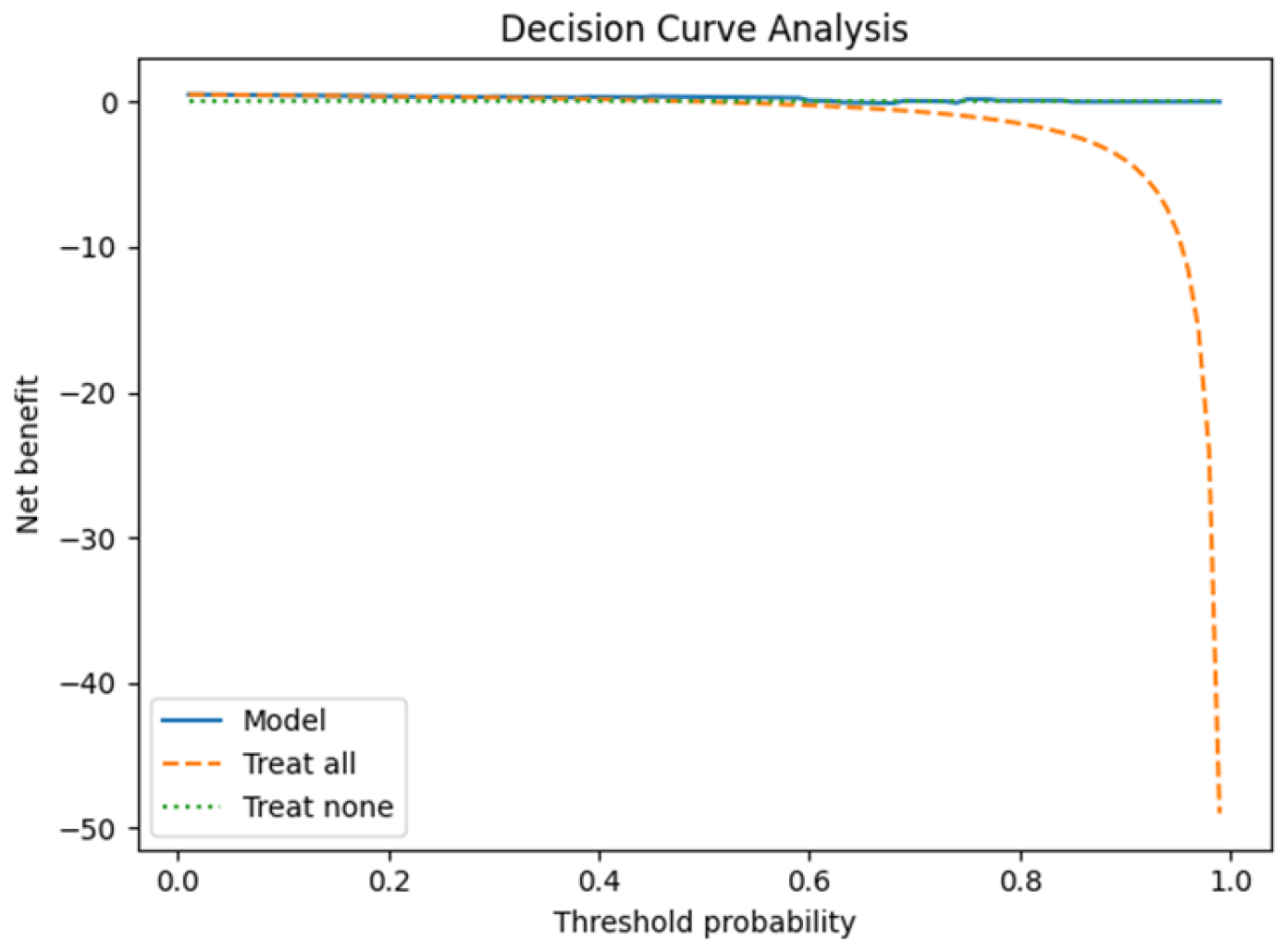
3.1. Leave-One-Patient-Out Cross-Validation (LOPO-CV)
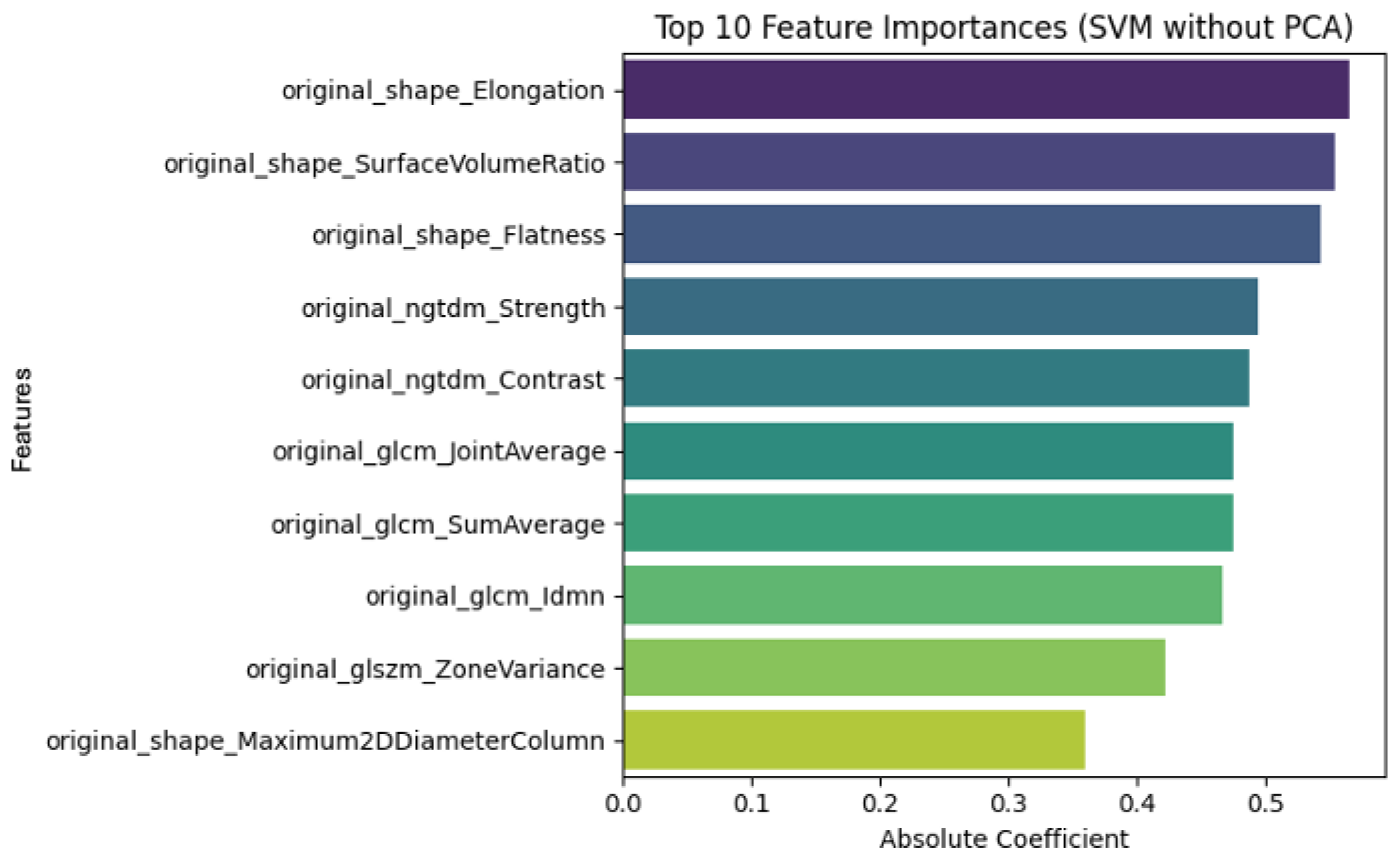
3.2. Radiomic Feature Interpretability and Clinical Relevance
4. Discussion
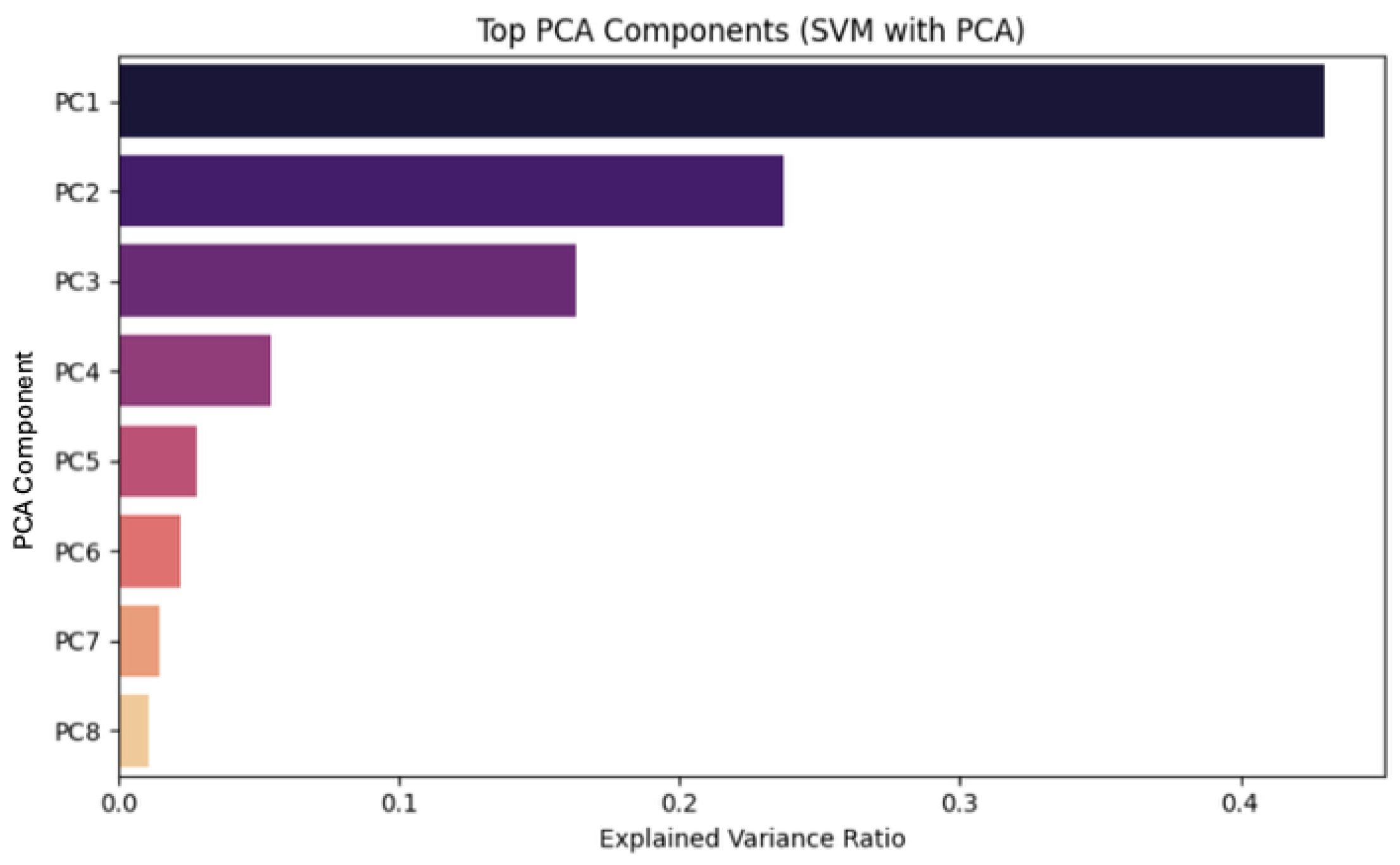
4.1. Dimensionality Reduction
4.2. SVM Classification and Model Robustness
4.3. Limitations and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BM | Brain Metastases |
| CNS | Central Nervous System |
| RT | Radiation Therapy |
| SRS | Stereotactic Radiosurgery |
| OARs | Organs at Risk |
| MRI | Magnetic Resonance Imaging |
| ROI | Region of Interest |
| PBC | Point-Biserial Correlation |
| CV | Cross-validation |
| AI | Artificial Intelligence |
| SVM | Support Vector Machine |
| PCA | Principal Component Analysis |
Appendix A
Appendix B
Appendix C
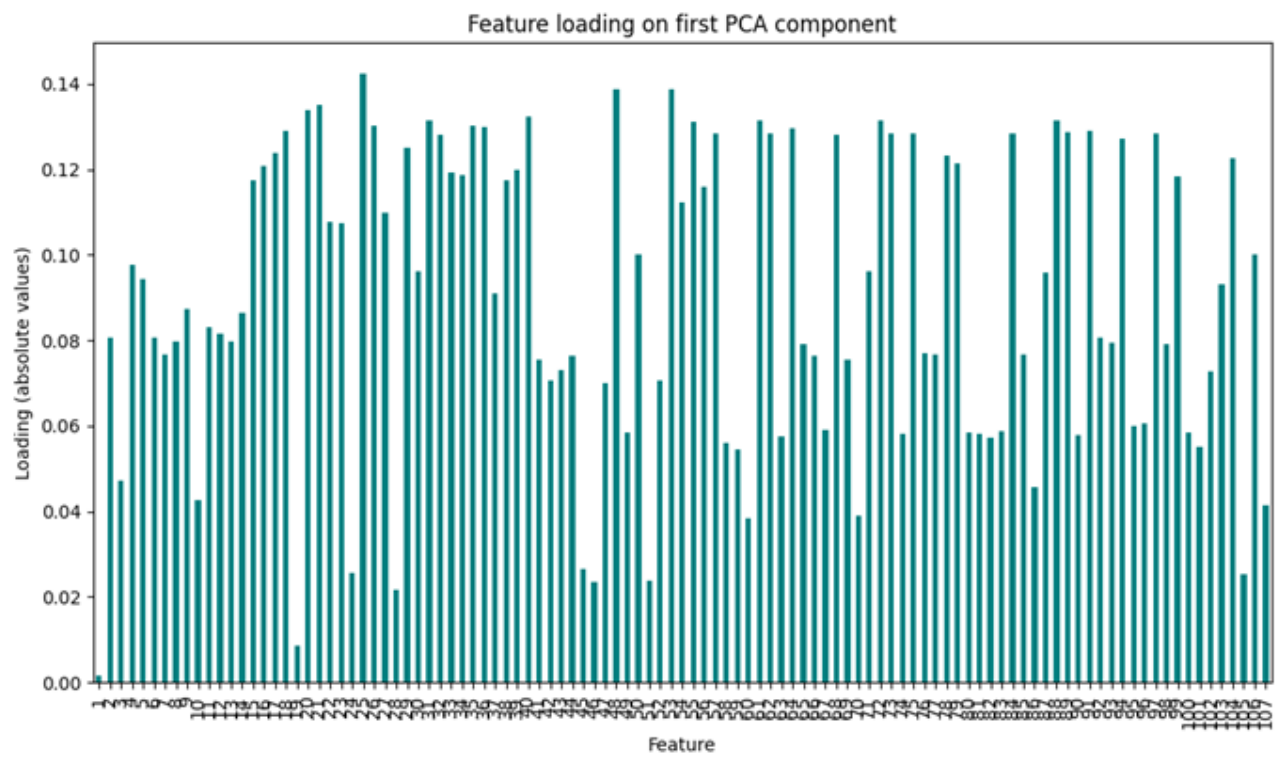
Appendix D
Appendix E
References
- Kotecha, R.; Gondi, V.; Ahluwalia, M.S.; Brastianos, P.K.; Mehta, M.P. Recent advances in managing brain metastasis. F1000Research 2018, 7, 1772. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rades, D.; Kieckebusch, S.; Haatanen, T.; Lohynska, R.; Dunst, J.; Schild, S.E. Surgical resection followed by whole brain radiotherapy versus whole brain radiotherapy alone for single brain metastasis. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, K.J. Epidemiology and prognosis of brain metastases. Surg. Neurol. Int. 2013, 4 (Suppl. S4), S192–S202. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cagney, D.N.; Martin, A.M.; Catalano, P.J.; Redig, A.J.; Lin, N.U.; Lee, E.Q.; Wen, P.Y.; Dunn, I.F.; Bi, W.L.; Weiss, S.E.; et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. Neurol. Oncol. 2017, 19, 1511–1521. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cross, N.E.; Glantz, M.J. Neurologic complications of radiation therapy. Neurol. Clin. 2003, 21, 249–277. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kawahara, D.; Tang, X.; Lee, C.K.; Nagata, Y.; Watanabe, Y. Predicting the local response of metastatic brain tumor to Gamma Knife radiosurgery by radiomics with a machine learning method. Front. Oncol. 2021, 10, 569461. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jajodia, A.; Goel, V.; Goyal, J.; Patnaik, N.; Khoda, J.; Pasricha, S.; Gairola, M. Combined Diagnostic Accuracy of Diffusion and Perfusion MR Imaging to Differentiate Radiation-Induced Necrosis from Recurrence in Glioblastoma. Diagnostics 2022, 12, 718. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- The MathWorks, Inc. MATLAB; vR2021b; The MathWorks, Inc.: Natick, MA, USA, 2021. [Google Scholar]
- Pasini, G.; Bini, F.; Russo, G.; Comelli, A.; Marinozzi, F.; Stefano, A. matRadiomics: A novel and complete radiomics framework, from image visualization to predictive model. J. Imaging 2022, 8, 221. [Google Scholar] [CrossRef] [PubMed]
- Python Software Foundation. Powerful Object-Oriented Programming Language; Python 3.12.8; Python Software Foundation: Wilmington, DE, USA, 2024; Available online: https://www.python.org/downloads/release/python-3128/ (accessed on 13 August 2025).
- Wang, Y.; Duggar, W.N.; Caballero, D.M.; Vengaloor Thomas, T.; Adari, N.; Mundra, E.K.; Wang, H. Brain tumor recurrence prediction after Gamma Knife radiotherapy from MRI and related DICOM-RT: An open annotated dataset and baseline algorithm (Brain-TR-GammaKnife) [Dataset]. Cancer Imaging Archive. 2023. Available online: https://www.cancerimagingarchive.net/collection/brain-tr-gammaknife/ (accessed on 24 August 2025). [CrossRef]




| Feature | Interpretation |
|---|---|
| Shape Features | |
| original_shape_Elongation | Measures how stretched the tumor is. A value close to 1 indicates a rounder shape; lower values indicate elongation. |
| original_shape_Flatness | Describes the flatness of the shape by assessing the ratio of the minor to major principal components. |
| original_shape_Maximum 2DDiameterColumn | Maximum 2D diameter of the lesion measured along the column direction. Useful for assessing size in a planar view. |
| original_shape_Sphericity | Quantifies how spherical the shape is. A value closer to 1 indicates a shape close to a perfect sphere. |
| First-order Statistics | |
| original_firstorder_Minimum | The lowest gray-level intensity value within the ROI. |
| original_firstorder_Skewness | Measures the asymmetry of the intensity distribution. High skewness indicates deviation from a normal distribution. |
| original_firstorder_Total_Energy | Sum of squared intensity values. Reflects the total signal magnitude within the region. |
| Texture Features (GLCM) | |
| original_glcm_Difference Entropy | Captures the randomness of intensity differences between neighboring voxels; higher values suggest greater heterogeneity. |
| original_glcm_Idmn | Inverse Difference Moment Normalized. Measures local homogeneity; higher values indicate more uniform texture. |
| original_glcm_Imc2 | Informational measure of correlation 2. Assess the complexity of the texture pattern. |
| Texture Features (GLSZM) | |
| original_glszm_ZoneVariance | Quantifies the variance in sizes of homogeneous zones; high values indicate varied and complex textures. |
| Texture Features (NGTDM) | |
| original_ngtdm_Contrast | Measures local contrast by comparing each voxel’s intensity with its neighborhood. |
| original_ngtdm_Strength | Represents the strength or coarseness of the perceived texture. |
| Ground Truth Label | |
| Gold_Standard | Binary label used as ground truth (e.g., 0 = control, 1 = incidence) for supervised learning. |
| Training Parameter | Values | Description |
|---|---|---|
| n_components | 0.90, 0.95, 0.99, None | Number of principal components to keep in PCA if it is an integer. Values between zero and one define the fraction of the total variance that should be preserved. |
| C | 0.1, 1, 10, 100 | Regularization parameter. Lower C allows wider margins with more tolerance to misclassification; higher C gives smaller margins and less tolerance to misclassification. The chosen range spans two orders of magnitude, thereby enabling the exploration of both under- and over-regularized regimes. |
| Kernel | linear, poly, rbf, sigmoid | Specifies the kernel type. In this work, all available kernels were used, i.e., linear, polynomial, radial basis function (rbf), and sigmoid. |
| Degree | 2, 3, 4 | Only used with a polynomial kernel, the degree defines the complexity of the feature mapping. Degrees 2 to 4 were selected to balance representational power with computational efficiency and to avoid excessive overfitting that might arise from higher-order polynomials. |
| gamma | auto, scale | Only used with Radial Basis Function and polynomial kernels, auto and scale constants provide two data-dependent normalizations without introducing arbitrary constant values. auto = 1/number of features scale = auto/variance of input data |
| #Metric | Mean Value | CI |
|---|---|---|
| Accuracy | 0.83 ± 0.11 | 0.58–1.00 |
| Sensitivity | 1.00 ± 0.00 | 1.00–1.00 |
| Specificity | 0.66 ± 0.21 | 0.20–1.00 |
| PPV | 0.75 ± 0.15 | 0.42–1.00 |
| NPV | 1.00 ± 0.00 | 1.00–1.00 |
| ROC-AUC | 0.80 ± 0.14 | 0.47–1.00 |
| PR-AUC | 0.83 ± 0.14 | 0.49–1.00 |
| Brier score | 0.18 ± 0.05 | – |
| PCA | C | Kernel |
|---|---|---|
| None | 0.1 | Linear |
| Feature Type | Feature Name | Clinical Insight | Patient Example | Confirmed Diagnosis |
|---|---|---|---|---|
| Texture | original_glcm_DifferenceEntropy | Highlights heterogeneity in necrotic tissue, useful for distinguishing from more homogeneous tumor recurrence. | GK_270 | Radiation necrosis |
| Shape | original_shape_Sphericity | Tumor recurrence tends to have more spherical shapes, while necrotic lesions are irregular due to tissue degradation. | GK_492 | Radiation necrosis |
| First-order | original_firstorder_Skewness | Reflects asymmetry in intensity, typical of necrotic tissue with edema and fibrosis. | GK_114 | Radiation necrosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frade, M.B.; Critelli, P.; Trifiletti, E.; Ripepi, G.; Pontoriero, A. Radiomic-Based Machine Learning for Differentiating Brain Metastases Recurrence from Radiation Necrosis Post-Gamma Knife Radiosurgery: A Feasibility Study. Int. J. Transl. Med. 2025, 5, 50. https://doi.org/10.3390/ijtm5040050
Frade MB, Critelli P, Trifiletti E, Ripepi G, Pontoriero A. Radiomic-Based Machine Learning for Differentiating Brain Metastases Recurrence from Radiation Necrosis Post-Gamma Knife Radiosurgery: A Feasibility Study. International Journal of Translational Medicine. 2025; 5(4):50. https://doi.org/10.3390/ijtm5040050
Chicago/Turabian StyleFrade, Mateus Blasques, Paola Critelli, Eleonora Trifiletti, Giuseppe Ripepi, and Antonio Pontoriero. 2025. "Radiomic-Based Machine Learning for Differentiating Brain Metastases Recurrence from Radiation Necrosis Post-Gamma Knife Radiosurgery: A Feasibility Study" International Journal of Translational Medicine 5, no. 4: 50. https://doi.org/10.3390/ijtm5040050
APA StyleFrade, M. B., Critelli, P., Trifiletti, E., Ripepi, G., & Pontoriero, A. (2025). Radiomic-Based Machine Learning for Differentiating Brain Metastases Recurrence from Radiation Necrosis Post-Gamma Knife Radiosurgery: A Feasibility Study. International Journal of Translational Medicine, 5(4), 50. https://doi.org/10.3390/ijtm5040050







