In Silico Identification of DNMT Inhibitors for the Treatment of Glioblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of Gene-Drug Interactions
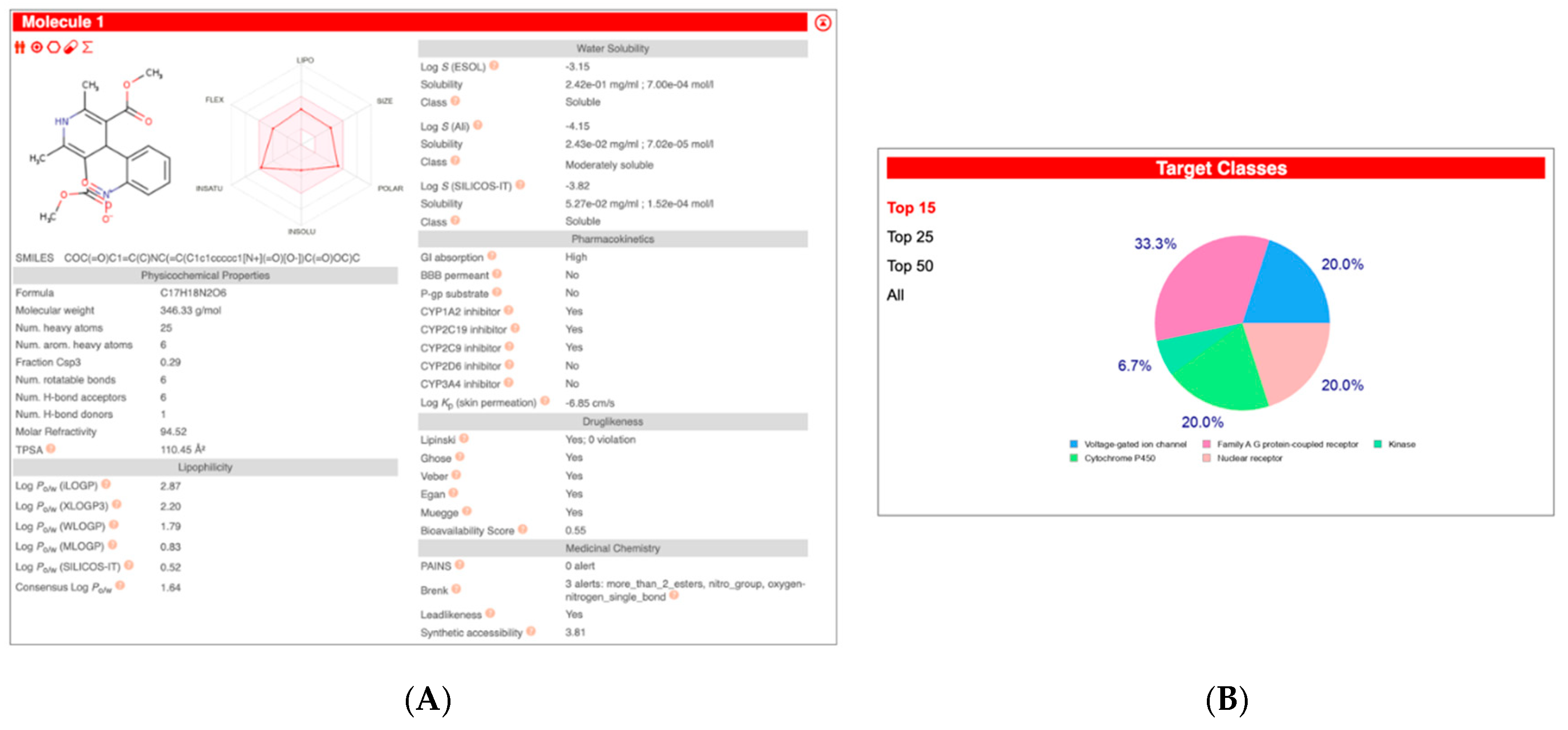
2.2. In Silico Analysis of Compounds
3. Result
3.1. The Responsiveness of Glioblastoma Cells to Potential Drugs
3.2. In Silico Draggability Analysis of Compounds
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNS | Central Nervous System |
| DNMTs | DNA Methyltransferases |
| DNMTis | DNA Methyltransferase Inhibitors |
| GBM | Glioblastoma Multiforme |
| FDA | The Food And Drug Administration |
| EMA | European Medicines Agency |
| MDS | Myelodysplastic Syndrome |
| AML | Acute Myeloid Leukaemia |
| CMML | Chronic Myelomonocytic Leukaemia |
| EGCG | Epigallocatechin |
| MCT KO | Monocrotaline Knockout |
| NIFE | Nifedipine |
References
- Kalkan, R. Epigenetics of Glioblastoma Multiforme. J. Clinic Res. Bioeth. 2015, 6, 225. Available online: https://www.omicsonline.org/open-access/epigenetics-of-glioblastoma-multiforme-2155-9627-1000225.php?aid=55742 (accessed on 3 July 2025).
- Kalkan, R. The Importance of Mutational Drivers in GBM. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Naser, A.Y.; Alwafi, H.; Hemmo, S.I.; Alrawashdeh, H.M.; Alqahtani, J.S.; Alghamdi, S.M.; Mustafa Ali, M.K. Trends in Hospital Admissions Due to Neoplasms in England and Wales between 1999 and 2019: An Ecological Study. Int. J. Environ. Res. Public Health 2022, 19, 8054. [Google Scholar] [CrossRef]
- Kampers, L.F.C.; Metselaar, D.S.; Vinci, M.; Scirocchi, F.; Veldhuijzen Van Zanten, S.; Eyrich, M.; Biassoni, V.; Hulleman, E.; Karremann, M.; Stücker, W.; et al. The Complexity of Malignant Glioma Treatment. Cancers 2025, 17, 879. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F. The Effect of 5-aza,2′-deoxyCytidine (5 AZA CdR or Decitabine) on Extrinsic, Intrinsic, and JAK/STAT Pathways in Neuroblastoma and Glioblastoma Cells Lines. Asian Pac. J. Cancer Prev. 2023, 24, 1841–1854. [Google Scholar] [CrossRef]
- Okuda, K.; Nakahara, K.; Ito, A.; Iijima, Y.; Nomura, R.; Kumar, A.; Fujikawa, K.; Adachi, K.; Shimada, Y.; Fujio, S.; et al. Pivotal role for S-nitrosylation of DNA methyltransferase 3B in epigenetic regulation of tumorigenesis. Nat. Commun. 2023, 14, 621. [Google Scholar] [CrossRef]
- Moshawih, S.; Lim, A.F.; Ardianto, C.; Goh, K.W.; Kifli, N.; Goh, H.P.; Jarrar, Q.; Ming, L.C. Target-Based Small Molecule Drug Discovery for Colorectal Cancer: A Review of Molecular Pathways and In Silico Studies. Biomolecules 2022, 12, 878. [Google Scholar] [CrossRef]
- Jan, Z.; Ahmed, W.S.; Biswas, K.H.; Jithesh, P.V. Identification of a potential DNA methyltransferase (DNMT) inhibitor. J. Biomol. Struct. Dyn. 2024, 42, 4730–4744. [Google Scholar] [CrossRef]
- Wang, K.; He, Z.; Jin, G.; Jin, S.; Du, Y.; Yuan, S.; Zhang, J. Targeting DNA methyltransferases for cancer therapy. Bioorganic Chem. 2024, 151, 107652. [Google Scholar] [CrossRef]
- Laranjeira, A.B.A.; Hollingshead, M.G.; Nguyen, D.; Kinders, R.J.; Doroshow, J.H.; Yang, S.X. DNA damage, demethylation and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci. Rep. 2023, 13, 5964. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, G.; Li, Y.; Lei, D.; Xiang, J.; Ouyang, L.; Wang, Y.; Yang, J. Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol. 2022, 13, 1072651. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zang, L.; Kondengaden, S.M.; Che, F.; Wang, L.; Heng, X. Potential Epigenetic-Based Therapeutic Targets for Glioma. Front. Mol. Neurosci. 2018, 11, 408. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; Van Den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, T.H.; Seo, W.S.; Yoo, S.D.; Kim, I.H.; Joo, S.H.; Shin, S.; Park, E.-S.; Ma, E.S.; Shin, B.S. Pharmacokinetics and tissue distribution of psammaplin A, a novel anticancer agent, in mice. Arch. Pharm. Res. 2012, 35, 1849–1854. [Google Scholar] [CrossRef] [PubMed]
- Wee, C.W.; Kim, J.H.; Kim, H.J.; Kang, H.C.; Suh, S.Y.; Shin, B.S.; Ma, E.; Kim, I.H. Radiosensitization of Glioblastoma Cells by a Novel DNA Methyltransferase-inhibiting Phthalimido-Alkanamide Derivative. Anticancer Res. 2019, 39, 759–769. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Chie, E.K.; Young, P.D.; Kim, I.A.; Kim, I.H. DNMT (DNA methyltransferase) inhibitors radiosensitize human cancer cells by suppressing DNA repair activity. Radiat. Oncol. 2012, 7, 39. [Google Scholar] [CrossRef]
- DepMap. DepMap 22Q2 Public. Figshare. 2022. Available online: https://figshare.com/articles/dataset/DepMap_22Q2_Public/19700056 (accessed on 3 July 2025).
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Napolitano, F.; Sirci, F.; Carrella, D.; Di Bernardo, D. Drug-set enrichment analysis: A novel tool to investigate drug mode of action. Bioinformatics 2016, 32, 235–241. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. Testing the predictive power of reverse screening to infer drug targets, with the help of machine learning. Commun. Chem. 2024, 7, 105. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef]
- Sengul, A.; Ünalan, S.; Can, S.; Gunay, O.; Karaçetin, D.; Sayyed, M.I. Volumetric rigid MR-CT registration for glioblastoma in radiation oncology: A Novel approach. J. Radiat. Res. Appl. Sci. 2024, 17, 100798. [Google Scholar] [CrossRef]
- Zabek, T.; Szmatola, T.; Adamczyk-Grochala, J.; Lewinska, A.; Wnuk, M. Knockout of TRDMT1 methyltransferase affects DNA methylome in glioblastoma cells. J. Neurooncol. 2023, 163, 61–69. [Google Scholar] [CrossRef]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Babaeenezhad, E.; Moradi Sarabi, M.; Rajabibazl, M.; Oraee-Yazdani, S.; Karima, S. Global and Regional DNA methylation silencing of PPARγ Associated with Glioblastoma Multiforme Pathogenesis. Mol. Biol. Rep. 2023, 50, 589–597. [Google Scholar] [CrossRef]
- Etcheverry, A.; Aubry, M.; De Tayrac, M.; Vauleon, E.; Boniface, R.; Guenot, F.; Saikali, S.; Hamlat, A.; Riffaud, L.; Menei, P. DNA methylation in glioblastoma: Impact on gene expression and clinical outcome. BMC Genom. 2010, 11, 701. [Google Scholar] [CrossRef]
- Fanelli, M.; Caprodossi, S.; Ricci-Vitiani, L.; Porcellini, A.; Tomassoni-Ardori, F.; Amatori, S.; Andreoni, F.; Magnani, M.; De Maria, R.; Santoni, A.; et al. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene 2008, 27, 358–365. [Google Scholar] [CrossRef]
- Purkait, S.; Sharma, V.; Kumar, A.; Pathak, P.; Mallick, S.; Jha, P.; Sharma, M.C.; Suri, V.; Julka, P.K.; Suri, A.; et al. Expression of DNA methyltransferases 1 and 3B correlates with EZH2 and this 3-marker epigenetic signature predicts outcome in glioblastomas. Exp. Mol. Pathol. 2016, 100, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Foltz, G.; Yoon, J.G.; Lee, H.; Ryken, T.C.; Sibenaller, Z.; Ehrich, M.; Hood, L.; Madan, A. DNA methyltransferase-mediated transcriptional silencing in malignant glioma: A combined whole-genome microarray and promoter array analysis. Oncogene 2009, 28, 2667–2677. [Google Scholar] [CrossRef] [PubMed]
- Hervouet, E.; Vallette, F.M.; Cartron, P.F. Impact of the DNA methyltransferases expression on the methylation status of apoptosis-associated genes in glioblastoma multiforme. Cell Death Dis. 2010, 1, e8. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Shi, Z.; Liu, X.; Zhang, A.; Han, L.; Jiang, K.; Kang, C.; Zhang, Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015, 359, 198–205. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, S.; Ibrahim, A.N.; Wang, J.; Deng, Z.; Wang, M. RCC2 promotes proliferation and radio-resistance in glioblastoma via activating transcription of DNMT1. Biochem. Biophys. Res. Commun. 2019, 516, 999–1006. [Google Scholar] [CrossRef]
- Lopez-Bertoni, H.; Lal, B.; Li, A.; Caplan, M.; Guerrero-Cázares, H.; Eberhart, C.G.; Quiñones-Hinojosa, A.; Glas, M.; Scheffler, B.; Laterra, J.; et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 2015, 34, 3994–4004. [Google Scholar] [CrossRef]
- Lv, Y.; Wang, C.; Liu, R.; Wu, S.; Chen, J.; Zheng, X.; Jiang, T.; Chen, L. NUP37 promotes the proliferation and invasion of glioma cells through DNMT1-mediated methylation. Cell Death Discov. 2024, 10, 373. [Google Scholar] [CrossRef]
- Li, Y.; Chen, F.; Chu, J.; Wu, C.; Li, Y.; Li, H.; Ma, H. miR-148-3p Inhibits Growth of Glioblastoma Targeting DNA Methyltransferase-1 (DNMT1). Oncol. Res. 2019, 27, 911–921. [Google Scholar] [CrossRef]
- Liao, C.H.; Lai, I.C.; Kuo, H.C.; Chuang, S.E.; Lee, H.L.; Whang-Peng, J.; Yao, C.-J.; Lai, G.-M. Epigenetic Modification and Differentiation Induction of Malignant Glioma Cells by Oligo-Fucoidan. Mar. Drugs 2019, 17, 525. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Z.; Ji, L.; Chen, C.; Wan, Q.; Xin, Y.; Pu, Z.; Li, K.; Jiao, J.; Yin, Y.; et al. SHF Acts as a Novel Tumor Suppressor in Glioblastoma Multiforme by Disrupting STAT3 Dimerization. Adv. Sci. 2022, 9, 2200169. [Google Scholar] [CrossRef]
- Abolfathi, S.; Zare, M. The evaluation of chitosan hydrogel based curcumin effect on DNMT1, DNMT3A, DNMT3B, MEG3, HOTAIR gene expression in glioblastoma cell line. Mol. Biol. Rep. 2023, 50, 5977–5989. [Google Scholar] [CrossRef]
- Xu, H.; Sun, J.; Shi, C.; Sun, C.; Yu, L.; Wen, Y.; Zhao, S.; Liu, J.; Xu, J.; Li, H.; et al. miR-29s inhibit the malignant behavior of U87MG glioblastoma cell line by targeting DNMT3A and 3B. Neurosci. Lett. 2015, 590, 40–46. [Google Scholar] [CrossRef]
- Apfel, C.C.; Korttila, K.; Abdalla, M.; Kerger, H.; Turan, A.; Vedder, I.; Zernak, C.; Danner, K.; Jokela, R.; Pocock, S.J.; et al. A Factorial Trial of Six Interventions for the Prevention of Postoperative Nausea and Vomiting. N. Engl. J. Med. 2004, 350, 2441–2451. [Google Scholar] [CrossRef]
- Chen, S.H.; Ke, T.L.; Shih, C.H.; Hsiung, C.N.; Chen, K.C.; Huang, Z.X.; Chuang, T.-H.; Chen, L.-K.; Chen, L. Reduced Taurine Synthesis Underlies Morphine-Promoted Lung Metastasis of Triple-Negative Breast Cancer. Cancers 2025, 17, 1086. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Yang, L.; Wu, H.; Xu, H.; Peng, J.; Wang, Y.; Zhou, F. Exploring the Molecular Mechanism of Comorbidity of Type 2 Diabetes Mellitus and Colorectal Cancer: Insights from Bulk Omics and Single-Cell Sequencing Validation. Biomolecules 2024, 14, 693. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, P.; Kaur, G.; Singh, N. Ozagrel a thromboxane A2 synthase inhibitor extenuates endothelial dysfunction, oxidative stress and neuroinflammation in rat model of bilateral common carotid artery occlusion induced vascular dementia. Vasc. Pharmacol. 2021, 137, 106827. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, P.; Singh, N. Thromboxane A2 synthase inhibition ameliorates endothelial dysfunction, memory deficits, oxidative stress and neuroinflammation in rat model of streptozotocin diabetes induced dementia. Physiol. Behav. 2021, 241, 113592. [Google Scholar] [CrossRef]
- Moussa, O.; Riker, J.M.; Klein, J.; Fraig, M.; Halushka, P.V.; Watson, D.K. Inhibition of thromboxane synthase activity modulates bladder cancer cell responses to chemotherapeutic agents. Oncogene 2008, 27, 55–62. [Google Scholar] [CrossRef]
- Machalz, D.; Li, H.; Du, W.; Sharma, S.; Liu, S.; Bureik, M.; Wolber, G. Discovery of a novel potent cytochrome P450 CYP4Z1 inhibitor. Eur. J. Med. Chem. 2021, 215, 113255. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, W.; Wu, X.; Shi, J. Pizotifen inhibits the proliferation and invasion of gastric cancer cells. Exp. Ther. Med. 2019, 19, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Piao, S.S.; Shang, B. Pizotifen Inhibits the Proliferation and Migration of Colon. Cancer HCT116 Cells by Down-regulating Wnt Signaling Pathway. Ann. Clin. Lab. Sci. 2019, 49, 183–188. [Google Scholar] [PubMed]
- Sarantos, M.R.; Papanikolaou, T.; Ellerby, L.M.; Hughes, R.E. Pizotifen Activates ERK and Provides Neuroprotection in vitro and in vivo in Models of Huntington’s Disease. J. Huntington Dis. 2012, 1, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Tan, L.; Li, Y.; Goh, B.C.; Wang, S.; Makinoshima, H.; Gong, Z. A zebrafish embryo screen utilizing gastrulation identifies the HTR2C inhibitor pizotifen as a suppressor of EMT-mediated metastasis. eLife 2021, 10, e70151, Erratum in eLife 2024, 13, e101706. [Google Scholar] [CrossRef]
- Wu, L.; Lin, W.; Liao, Q.; Wang, H.; Lin, C.; Tang, L.; Lian, W.; Chen, Z.; Li, K.; Xu, L. Calcium Channel Blocker Nifedipine Suppresses Colorectal Cancer Progression and Immune Escape by Preventing NFAT2 Nuclear Translocation. Cell Rep. 2020, 33, 108327. [Google Scholar] [CrossRef]
- Kondo, S.; Yin, D.; Morimura, T.; Kubo, H.; Nakatsu, S.; Takeuchi, J. Combination therapy with cisplatin and nifedipine induces apoptosis in cisplatin-sensitive and cisplatin-resistant human glioblastoma cells. Br. J. Cancer 1995, 71, 282–289. [Google Scholar] [CrossRef]
- Chen, J.W.; Madamanchi, N.; Madamanchi, N.R.; Trier, T.T.; Keherly, M.J. Lamp-1 is upregulated in human glioblastoma cell lines induced to undergo apoptosis. J. Biomed. Sci. 2001, 8, 365–374. [Google Scholar] [CrossRef]
- Varricchio, A.; Khan, S.; Price, Z.K.; Davis, R.A.; Ramesh, S.A.; Yool, A.J. Pharmacological Inhibition of Membrane Signaling Mechanisms Reduces the Invasiveness of U87-MG and U251-MG Glioblastoma Cells In Vitro. Cancers 2023, 15, 1027. [Google Scholar] [CrossRef]
- Tang, W.; Zhu, X.; Bian, L.; Zhang, B. Research progress of dydrogesterone in the treatment of endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2024, 296, 120–125. [Google Scholar] [CrossRef]
- Peng, C.; Huang, Y.; Zhou, Y. Dydrogesterone in the treatment of endometriosis: Evidence mapping and meta-analysis. Arch. Gynecol. Obstet. 2021, 304, 231–252. [Google Scholar] [CrossRef]
- Piccinni, M.P.; Raghupathy, R.; Saito, S.; Szekeres-Bartho, J. Cytokines, Hormones and Cellular Regulatory Mechanisms Favoring Successful Reproduction. Front. Immunol. 2021, 12, 717808. [Google Scholar] [CrossRef]
- Dias-Neto, M.; Luísa-Neves, A.; Pinho, S.; Gonçalves, N.; Mendes, M.; Eloy, C.; Lopes, J.M.; Gonçalves, D.; Ferreira-Pinto, M.; Leite-Moreira, A.F. Pathophysiology of Infantile Pulmonary Arterial Hypertension Induced by Monocrotaline. Pediatr. Cardiol. 2015, 36, 1000–1013. [Google Scholar] [CrossRef]
- Wang, Q.; Chai, L.; Zhang, Q.; Wang, J.; Liu, J.; Chen, H.; Wang, Y.; Chen, Y.; Shen, N.; Xie, X.; et al. Induction of GLI1 by miR-27b-3p/FBXW7/KLF5 pathway contributes to pulmonary arterial hypertension. J. Mol. Cell Cardiol. 2022, 171, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Bi, R.; Bao, C.; Jiang, L.; Liu, H.; Yang, Y.; Mei, J.; Ding, F. MicroRNA-27b plays a role in pulmonary arterial hypertension by modulating peroxisome proliferator-activated receptor γ dependent Hsp90-eNOS signaling and nitric oxide production. Biochem. Biophys. Res. Commun. 2015, 460, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Zhang, S.; Wang, X.; Ran, G.; Zhang, X.; Xi, J.; Gao, Z.; Lei, Y.; Pan, J.; Liu, Y.; et al. Inflammation Intensifies Monocrotaline-Induced Liver Injury. J. Agric. Food Chem. 2023, 71, 3433–3443. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yuan, Y.; Wang, R.; Bai, J.; Jia, Y.; Qiu, X.; Niu, H.; Li, L.; Luo, Y.; Zhao, B.; et al. Monocrotaline-mediated autophagy via inhibiting PI3K/AKT/mTOR pathway induces apoptosis in rat hepatocytes. Front. Pharmacol. 2024, 15, 1499116. [Google Scholar] [CrossRef]
- Kusuma, S.S.; Tanneeru, K.; Didla, S.; Devendra, B.N.; Kiranmayi, P. Antineoplastic activity of monocrotaline against hepatocellular carcinoma. Anticancer. Agents Med. Chem. 2014, 14, 1237–1248. [Google Scholar] [CrossRef]
- Alqumaya, A.S.; Alresheedi, T.; Al-Yusufy, F.; El-Sayed, W.A.; Aroua, L. Pyrazole Derivatives: A New Synthesis, Biological Importance, and Recent Practical Applications. J. Qassim Univ. Sci. 2025, 4, 25–73. [Google Scholar]
- Horwitz, A.B.; Rubin, R.T. Barbiturates and pyrazolopyridines for the treatment of postpartum depression-repurposing of two drug classes. Front Pharmacol. 2023, 14, 1139889. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692.73. [Google Scholar] [CrossRef]
- Tsai, H.C. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012, 21, 430–446. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and clinical applications. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef]
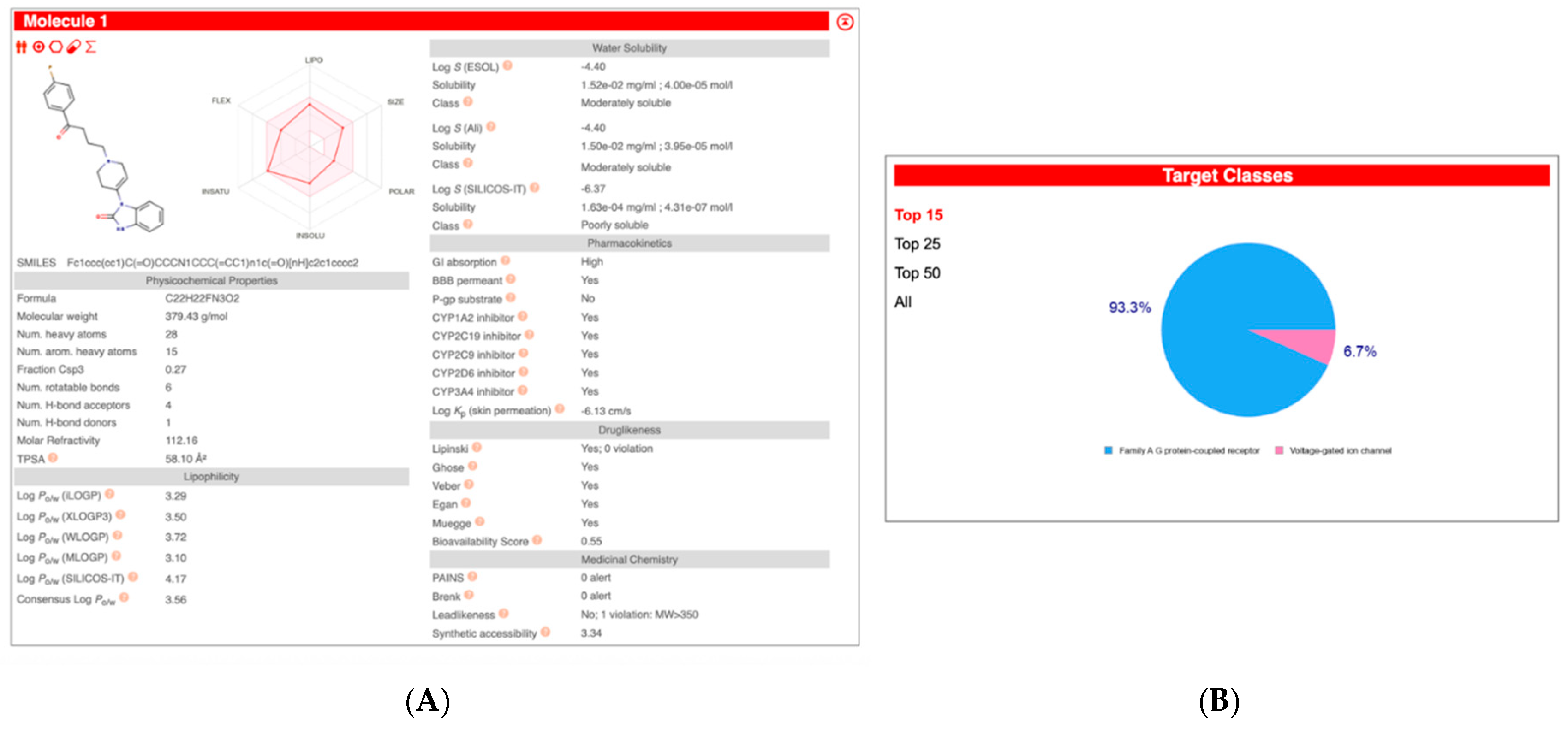

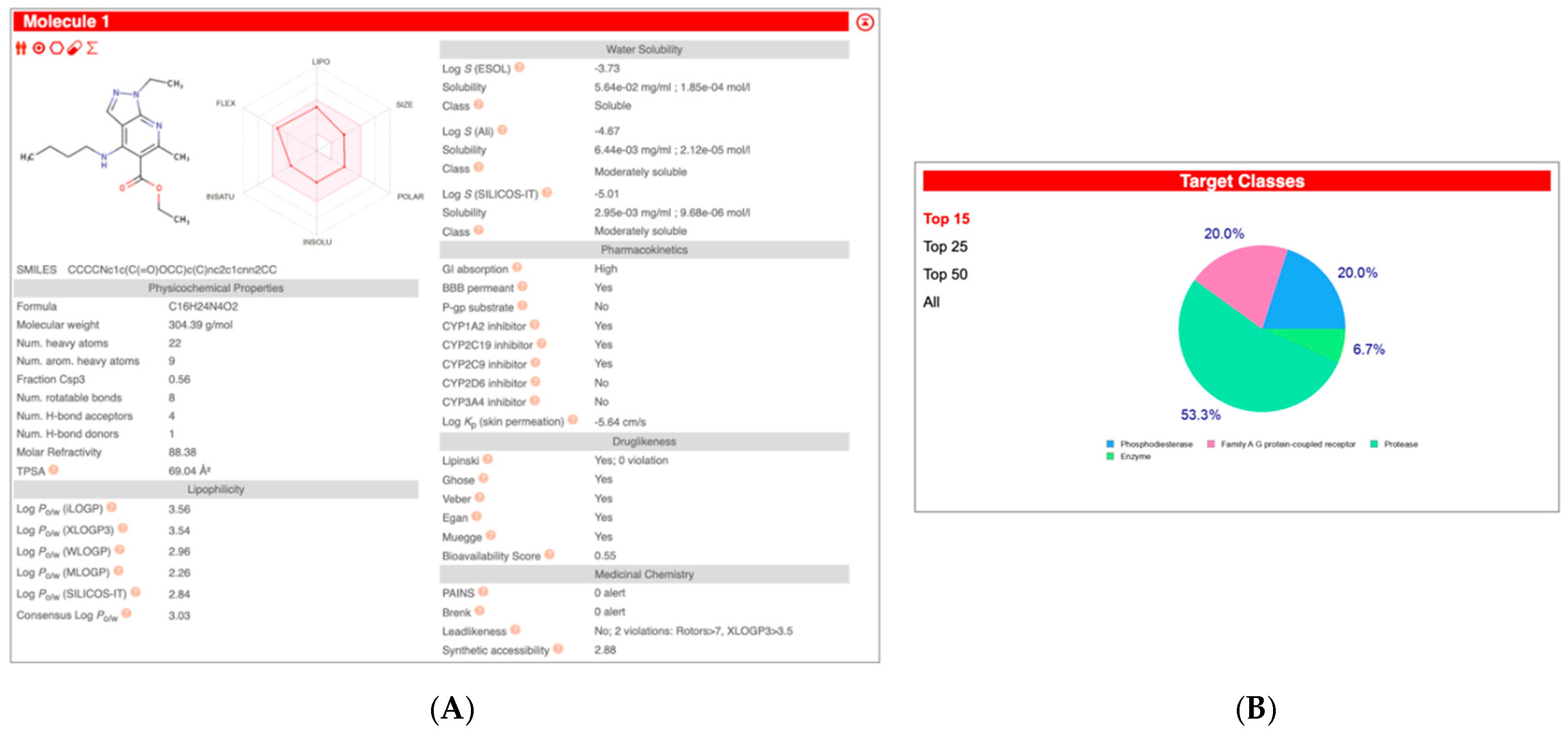
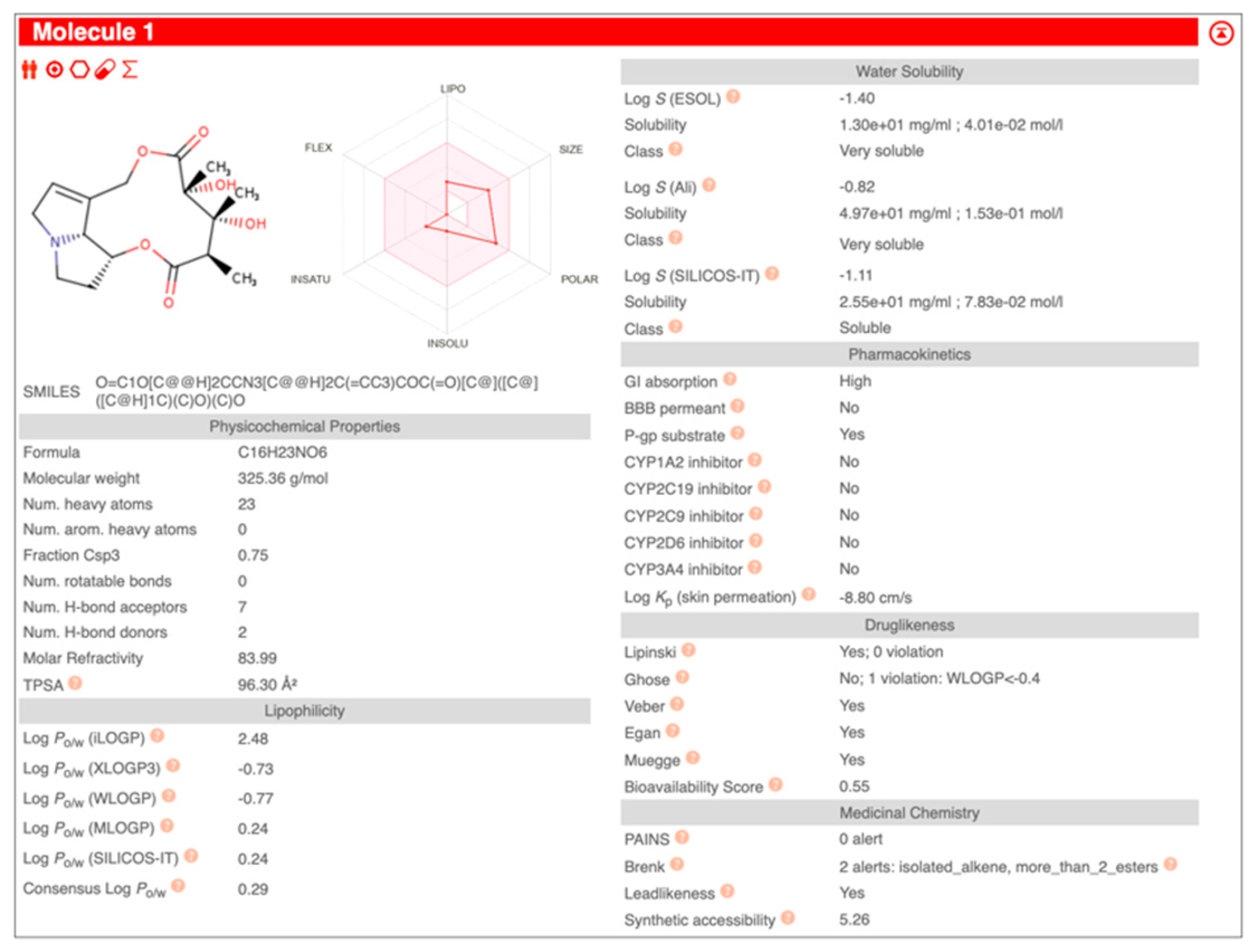
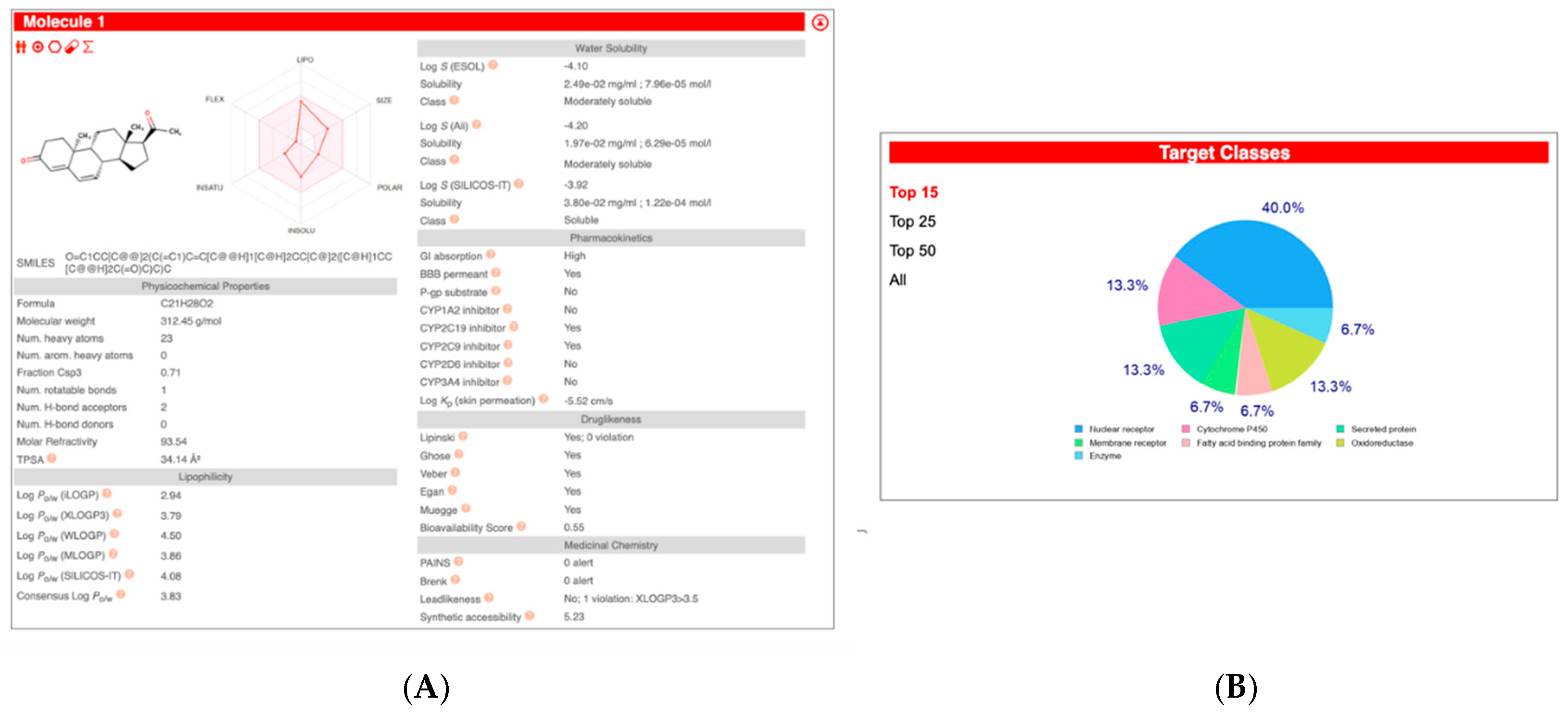
| Compound Name | Pearson | Spearman | Slope | Intercept | p-Value (Linregress) |
|---|---|---|---|---|---|
| droperidol | −0.470 | −0.535 | −2.75 × 10−1 | 9.11 × 10−1 | 8.81 × 10−3 |
| demeclocycline | −0.436 | −0.401 | −2.53 × 10−1 | 6.16 × 10−1 | 5.54 × 10−3 |
| benzthiazide | −0.465 | −0.365 | −2.33 × 10−1 | 2.01 × 10−1 | 9.54 × 10−3 |
| ozagrel | 0.529 | 0.515 | 2.28 × 10−1 | −6.23 × 10−1 | 3.77 × 10−3 |
| pizotifen | 0.590 | 0.605 | 2.71 × 10−1 | −9.04 × 10−1 | 7.53 × 10−4 |
| Compound Name | Pearson | Spearman |
|---|---|---|
| tracazolate | −0.497 | −0.522 |
| vorinostat:navitoclax (4:1) | 0.486 | 0.490 |
| norcyclobenzaprine | −0.494 | −0.447 |
| Compound Name | Pearson | Spearman | Slope | Intercept | p-Value (Linregress) |
|---|---|---|---|---|---|
| tracazolate | −0.507 | −0.485 | −3.63 × 10−1 | 4.98 × 10−1 | 4.98 × 10−3 |
| monocrotaline | 0.454 | 0.310 | 2.65 × 10−1 | −7.41 × 10−1 | 3.72 × 10−3 |
| dydrogesterone | 0.525 | 0.614 | 3.22 × 10−1 | −3.98 × 10−1 | 2.88 × 10−3 |
| 6-benzylaminopurine | −0.571 | −0.567 | −2.72 × 10−1 | 4.77 × 10−1 | 1.21 × 10−3 |
| nifedipine | −0.497 | −0.573 | −4.40 × 10−1 | 7.14 × 10−1 | 1.29 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osum, M.; Alsaloumi, L.; Kalkan, R. In Silico Identification of DNMT Inhibitors for the Treatment of Glioblastoma. Int. J. Transl. Med. 2025, 5, 48. https://doi.org/10.3390/ijtm5040048
Osum M, Alsaloumi L, Kalkan R. In Silico Identification of DNMT Inhibitors for the Treatment of Glioblastoma. International Journal of Translational Medicine. 2025; 5(4):48. https://doi.org/10.3390/ijtm5040048
Chicago/Turabian StyleOsum, Meyrem, Louai Alsaloumi, and Rasime Kalkan. 2025. "In Silico Identification of DNMT Inhibitors for the Treatment of Glioblastoma" International Journal of Translational Medicine 5, no. 4: 48. https://doi.org/10.3390/ijtm5040048
APA StyleOsum, M., Alsaloumi, L., & Kalkan, R. (2025). In Silico Identification of DNMT Inhibitors for the Treatment of Glioblastoma. International Journal of Translational Medicine, 5(4), 48. https://doi.org/10.3390/ijtm5040048






