Pseudomonas aeruginosa Biofilm Formation and Its Control
Abstract
1. Introduction
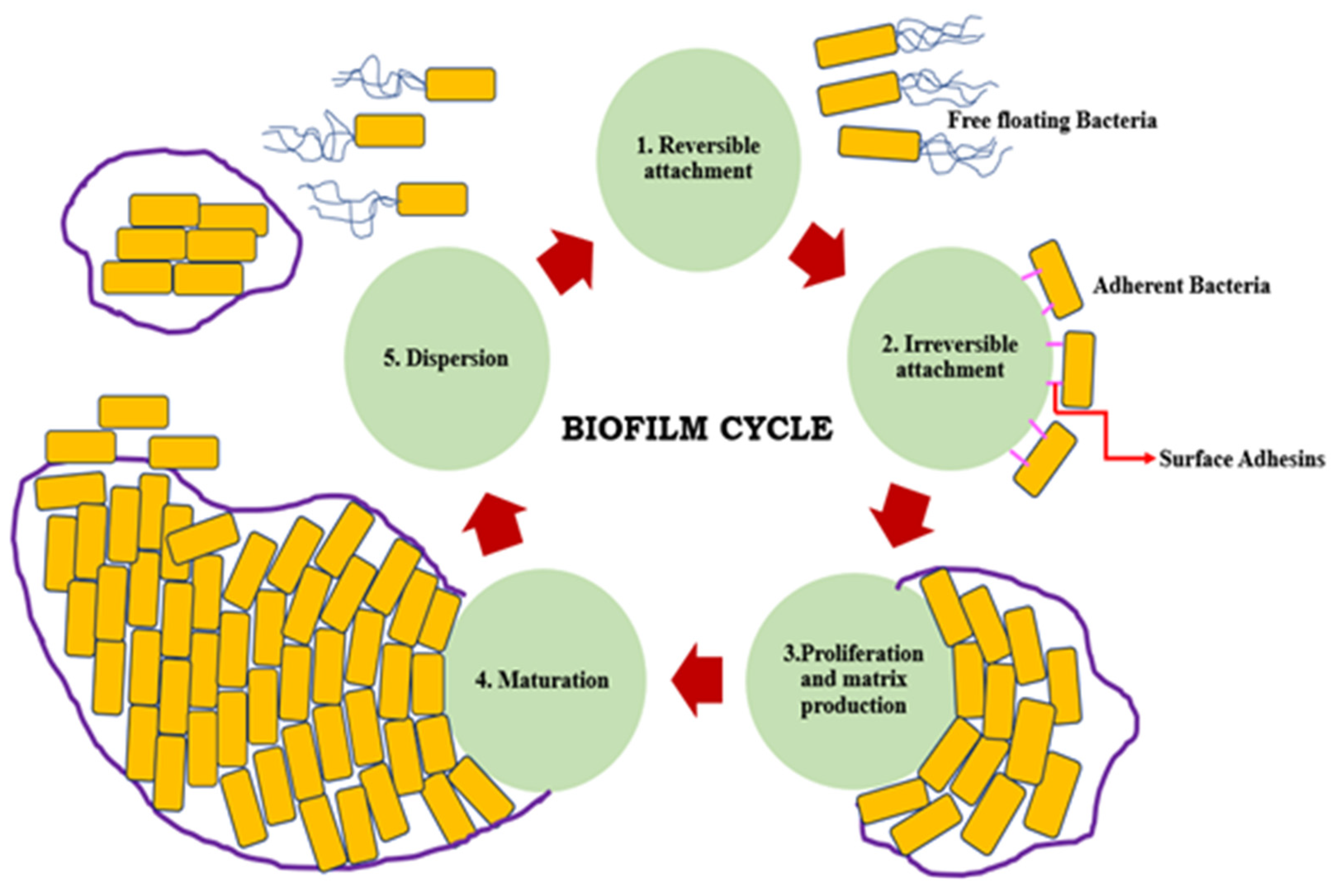
2. Formation of Biofilm
- Adherence to each other
- Adherence to either solid/liquid, solid/air, liquid/liquid, or liquid/air interfaces
- Attachment to surfaces
- Decreased antimicrobial susceptibility
- Decreased host defense systems
- Existence of one or more microbial species
- Three-dimensional structure
3. Pseudomonas aeruginosa Biofilms
4. Role of Quorum Sensing (QS) in Biofilm Formation
5. Pseudomonas aeruginosa Quorum Sensing System
6. Pseudomonas aeruginosa Biofilm Challenge to Antimicrobial Agents
7. Strategies to Control P. aeruginosa Biofilm Infections
7.1. Plants as a Natural Source of Antibiofilm Agents for P. aeruginosa Biofilms
7.2. Enzymes against P. aeruginosa Biofilms
7.3. In Silico Approach to Control P. aeruginosa Biofilms
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010, 59, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lee, J. Antibiofilm agents: A new perspective for antimicrobial strategy. J. Microbiol. 2017, 55, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle:A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Prakash, B.; Veeregowda, B.M.; Krishnappa, G. Biofilms: A survival strategy of bacteria. Curr. Sci. 2003, 85, 1299–1307. [Google Scholar]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Häussler, S.; Parsek, M.R. Biofilms 2009: New perspectives at the heart of surface-associated microbial communities. J. Bacteriol. 2010, 192, 2941–2949. [Google Scholar] [CrossRef]
- Schwartz, T.; Jungfer, C.; Heisler, S.; Friedrich, F.; Faubel, W.; Obst, U. Combined use of molecular biology taxonomy, Raman spectrometry, and ESEM imaging to study natural biofilms grown on filter materials at waterworks. Chemosphere 2009, 77, 249–257. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.; Ryu, S.Y.; Cho, M.H.; Lee, J. Ginkgolic acids and Gingko biloba extract inhibit Escherichia coli O157: H7 and Staphylococcus aureus biofilm formation. Int. J. Food Microbiol. 2014, 174, 47–55. [Google Scholar] [CrossRef]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Wu, H.; Hóiby, N.; Molin, S.; Song, Z. Current understanding of multi species biofilms. Int. J. Oral Sci. 2011, 3, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Briedenstein, E.B.M.; Hancock, R.E.W. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updates 2011, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Francolini, I.; Donelli, G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 2010, 59, 227–238. [Google Scholar] [CrossRef]
- Mermel, L.A.; Farr, B.M.; Sherertz, R.J.; Raad, I.I.; O’ Grady, N.; Harris, J.S.; Craven, D.E. Guidelines for the management of intravascular catheter-related infections. Clin. Infect. Dis. 2001, 32, 1249–1272. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M. Invasive methicillin-resistant Staphylococcus aureus infections in United States. JAMA 2007, 298, 1763–1771. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.G.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Genet. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Pinto, H.; Simões, M.; Borges, A. Prevalence and impact of biofilms on bloodstream and urinary tract infections:A systematic review and meta-analysis. Antibiotics 2021, 10, 825. [Google Scholar] [CrossRef]
- Wenzel, R.P.; Edmond, M.B. The impact of hospital-acquired bloodstream infections. Emerg. Infect. Dis. 2001, 7, 174–177. [Google Scholar] [CrossRef]
- Bryers, J.D. Medical biofilms. Biotechnol. Bioeng. 2008, 100, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, R.P. Health care-associated infections: Major issues in the early years of the 21st century. Clin. Infect. Dis. 2007, 45 (Suppl. 1), S85–S88. [Google Scholar] [CrossRef] [PubMed]
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Maukonen, J.; Mättö, J.; Wirtanen, G.; Raaska, L.; Mattila-Sandholm, T.; Saarela, M. Methodologies for the characterization of microbes in industrial environments: A review. J. Ind. Microbiol. Biotechnol. 2003, 30, 327–356. [Google Scholar] [CrossRef] [PubMed]
- Sihorkar, V.; Vyas, S.P. Biofilm consortia on biomedical and biological surfaces: Delivery and targeting strategies. Pharm. Res. 2001, 18, 1247–1254. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Bjarnsholt, T.; Jensen, P.Ø.; Givskov, M.; Høiby, N. Targeting quorum sensing in Pseudomonas aeruginosa biofilms: Current and emerging inhibitors. Future Microbiol. 2013, 8, 901–921. [Google Scholar] [CrossRef]
- Rahim, K.; Saleha, S.; Basit, A.; Zhu, X.; Ahmed, I.; Huo, L.; Zhang, P.; Usman, B.; Munir, S.; Franco, O.L. Pseudomonas aeruginos as a powerful biofilm producer and positive action of amikacin against isolates from chronic wounds. Jundishapur. J. Microbiol. 2017, 10, e57564. [Google Scholar] [CrossRef]
- Wojtyczka, R.D.; Orlewska, K.; Kępa, M.; Idzik, D.; Dziedzic, A.; Mularz, T.; Krawczyk, M.; Miklasińska, M.; Wąsik, T.J. Biofilm formation and antimicrobial susceptibility of Staphylococcus epidermidis strains from a hospital environment. Int. J. Environ. Res. Public Health 2014, 11, 4619–4633. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J. Management of biofilm-associated infections: What can we expect from recent research on biofilm lifestyles? Med. Sci. 2012, 28, 727–739. [Google Scholar]
- Lerch, T.Z.; Chenu, C.; Dignac, M.F.; Barriuso, E.; Mariotti, A. Biofilm vs. Planktonic lifestyle: Consequences for pesticide 2,4-D metabolism by Cupriavidus necator. Front. Microbiol. 2017, 8, 904. [Google Scholar] [CrossRef]
- McCarty, S.; Woods, E.; Percival, S.L. Biofilms: From Concept to Reality; Elsevier Inc.: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Fagerlind, M.G.; Webb, J.S.; Barraud, N.; McDougald, D.; Jansson, A.; Nilsson, P.; Harlén, M.; Kjelleberg, S.; Rice, S.A. Dynamic modelling of cell death during biofilm development. J. Theor. Biol. 2012, 295, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Olivares, E.; Badel-Berchoux, S.; Provot, C.; Prévost, G.; Bernardi, T.; Jehl, F. Clinical impact of antibiotics for the treatment of Pseudomonas aeruginosa biofilm infections. Front. Microbiol. 2020, 10, 2894. [Google Scholar] [CrossRef] [PubMed]
- Dufour, D.; Leung, V.; Lévesque, C.M. Bacterial biofilm: Structure, function, and antimicrobial resistance. Endod. Topics 2010, 22, 2–16. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Annous, B.A.; Fratamico, P.M.; Smith, J.L. Quorum sensing in biofilms: Why bacteria behave the way they do. J. Food Sci. 2009, 74, 24–37. [Google Scholar] [CrossRef]
- Flemming, H.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef]
- Hausner, M.; Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 1999, 65, 3710–3713. [Google Scholar] [CrossRef]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTierman, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, 201067. [Google Scholar] [CrossRef]
- Srivastava, S.; Bhargava, A. Biofilms and human health. Biotechnol. Lett. 2016, 38, 1–22. [Google Scholar] [CrossRef]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Bonaventura, G.D.; Hébraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef]
- Turan, O.; Deirel, Y.; Day, S.; Tezdogan, T. Experimental determination of added hydrodynamic resistance caused by marine biofouling on ships. In Proceedings of the 6th European Transport Research Conference, Warsaw, Poland, 18–21 April 2016; pp. 1–10. [Google Scholar]
- Abdel-Aziz, S.M.; Aeron, A. Bacterial biofilm: Dispersal and inhibition strategies. SAJ Biotechnol. 2014, 1, 105. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Q.; Sun, H. Novel strategies for the prevention and treatment of biofilm related infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y. Surface sensing for biofilm formation in Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 2671. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Schneper, L.; Kumari, H.; Mathee, K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013, 41, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Streeter, K.; Katouli, M. Pseudomonas aeruginosa: A review of their pathogenesis and prevalence in clinical settings and the environment. Infect. Epidemiol. Med. 2016, 2, 25–32. [Google Scholar] [CrossRef]
- Mantero, M.; Gramegna, A.; Pizzamiglio, G.; D’Adda, A.; Tarsia, P.; Blasi, F. Once daily aerosolised tobramycin in adult patients with cystic fibrosis in the management of Pseudomonas aeruginosa chronic infection. Multidiscip. Respir. Med. 2017, 12, 2. [Google Scholar] [CrossRef]
- Davis, R.; Brown, P.D. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J. Med. Microbiol. 2016, 65, 261–271. [Google Scholar] [CrossRef]
- Valderrey, A.D.; Pozuelo, M.J.; Jiménez, P.A.; Maciá, M.D.; Oliver, A.; Rotger, R. Chronic colonization by Pseudomonas aeruginosa of patients with obstructive lung diseases: Cystic fibrosis, bronchiectasis, and chronic obstructive pulmonary disease. Diagn. Microbiol. Infect. Dis. 2010, 68, 20–27. [Google Scholar] [CrossRef]
- Kerr, K.G.; Snelling, A.M. Pseudomonas aeruginosa: A formidable and ever-present adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K.; National Healthcare Safety Network Team; Participating National Healthcare Safety Network Facilities. NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare associated infections: Annual summary of data reported to the National Healthcare Safety Network at the centers for disease control and prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef]
- Strateva, T.; Mitov, I. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann. Microbiol. 2011, 61, 717–732. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H.A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef] [PubMed]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015, 759348, 759348. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wang, J.; Wang, S.; Anderson, E.M.; Lam, J.S.; Parsek, M.R.; Wozniak, D.J. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post transcriptionally regulated. Environ. Microbiol. 2012, 14, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.L.; Parsek, M.R. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011, 7, e1001264. [Google Scholar] [CrossRef]
- Franklin, M.J.; Nivens, D.E.; Weadge, J.T.; Howell, P.L. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2011, 167, 1–16. [Google Scholar] [CrossRef]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 2010, 12, 1621–1629. [Google Scholar]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.T.; Matewish, J.M.; Kessler, J.L.; Hyodo, M.; Hayakawa, Y.; Lory, S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 2007, 65, 1474–1484. [Google Scholar] [CrossRef]
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analysis of Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Regni, C.; Tipton, P.A.; Beamer, L.J. Crystal structure of PMM/PGM: An enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 2002, 10, 269–279. [Google Scholar] [CrossRef]
- Fritz, T.A.; Hurley, J.H.; Trinh, L.B.; Shiloach, J.; Tabak, L.A. The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide alpha-Nacetylgalactosaminyltransferase- T1. Proc. Natl. Acad. Sci. USA 2004, 101, 15307–15312. [Google Scholar] [CrossRef] [PubMed]
- Ramelot, T.A.; Yee, A.; Cort, J.R.; Semesi, A.; Arrowsmith, C.H.; Kennedy, M.A. NMR structure and binding studies confirmthat PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 2007, 66, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Keiski, C.L.; Harwich, M.; Jain, S.; Neculai, A.M.; Yip, P.; Robinson, H.; Whitney, J.C.; Riley, L.; Burrows, L.L.; Ohman, D.E.; et al. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 2010, 18, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Rozeboom, H.J.; Bjerkan, T.M.; Kalk, K.H.; Ertesvag, H.; Holtan, S.; Aachmann, F.L.; Valla, S.; Dijkstra, B.W. Structural and mutational characterization of the catalytic A-module of the mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii. J. Biol. Chem. 2008, 283, 23819–23828. [Google Scholar] [CrossRef]
- Molgaard, A.; Larsen, S. A branched N-linked glycan at atomic resolution in the 1.12 A structure of rhamnogalacturonan acetylesterase. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 A resolution. J. Mol. Biol. 1999, 290, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Franklin, M.J.; Ohman, D.E. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 3000–3007. [Google Scholar] [CrossRef] [PubMed]
- Pelissier, M.C.; Lesley, S.A.; Kuhn, P.; Bourne, Y. Structural insights into the catalytic mechanism of bacterial guanosinediphospho- D-mannose pyrophosphorylase and its regulation by divalent ions. J. Biol. Chem. 2010, 285, 27468–27476. [Google Scholar] [CrossRef] [PubMed]
- Urch, J.E.; Hurtado-Guerrero, R.; Brosson, D.; Liu, Z.; Eijsink, V.G.; Texier, C.; Van Aalten, D.M. Structural and functional characterization of a putative polysaccharide deacetylase of the human parasite Encephalitozoon cuniculi. Protein Sci. 2009, 18, 1197–1209. [Google Scholar] [CrossRef]
- Zhang, Z.; Kulkarni, K.; Hanrahan, S.J.; Thompson, A.J.; Barford, D. The APC/C subunit Cdc16/Cut9 is a contiguous tetratricopeptide repeat superhelix with a homo-dimer interface similar to Cdc27. EMBO J. 2010, 29, 3733–3744. [Google Scholar] [CrossRef]
- Bonsor, D.A.; Grishkovskaya, I.; Dodson, E.J.; Kleanthous, C. Molecular mimicry enables competitive recruitment by a natively disordered protein. J. Am. Chem. Soc. 2007, 129, 4800–4807. [Google Scholar] [CrossRef]
- Wang, H.; Robinson, H.; Ke, H. Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. J. Biol. Chem. 2010, 285, 38149–38156. [Google Scholar] [CrossRef]
- Li, J.; Qian, X.; Hu, J.; Sha, B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 2009, 284, 23852–23859. [Google Scholar] [CrossRef]
- Chua, T.K.; Bujnicki, J.M.; Tan, T.C.; Huynh, F.; Patel, B.K.; Sivaraman, J. The structure of sucrose phosphate synthase from Halothermothrix orenii reveals its mechanism of action and binding mode. Plant Cell 2008, 20, 1059–1072. [Google Scholar] [CrossRef]
- He, X.; Szewczyk, P.; Karyakin, A.; Evin, M.; Hong, W.X.; Zhang, Q.; Chang, G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 2010, 467, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Beis, K.; Nesper, J.; Brunkan- Lamontagne, A.L.; Clarke, B.R.; Whitfield, C.; Naismith, J.H. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 2006, 444, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Bechet, E.; Gruszczyk, J.; Terreux, R.; Gueguen-Chaignon, V.; Vigouroux, A.; Obadia, B.; Cozzone, A.J.; Nessler, S.; Grangeasse, C. Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol. Microbiol. 2010, 77, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.E.; Kordulakova, J.; Schaeffer, F.; Svetlikova, Z.; Buschiazzo, A.; Giganti, D.; Gicquel, B.; Mikusova, K.; Jackson, M.; Alzari, P.M. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyl transferase PimA from mycobacteria. J. Biol. Chem. 2007, 282, 20705–20714. [Google Scholar] [CrossRef]
- Yang, J.K.; Yoon, H.J.; Ahn, H.J.; Lee, B.I.; Pedelacq, J.D.; Liong, E.C.; Berendzen, J.; Laivenieks, M.; Vieille, C.; Zeikus, G.J.; et al. Crystal structure of beta-Dxylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase. J. Mol. Biol. 2004, 335, 155–165. [Google Scholar] [CrossRef]
- Favre-Bonté, S.; Chamot, E.; Köhler, T.; Romand, J.; van Delden, C. Auto inducer production and quorum-sensing dependent phenotypes of Pseudomonas aeruginosa vary according to isolation site during colonization of incubated patients. BMC Microbiol. 2007, 7, 33. [Google Scholar] [CrossRef]
- Serralta, V.W.; Harison-Balestra, C.; Cazzaniga, A.L.; Davis, S.C.; Mertz, P.M. Lifestyles of bacteria in wounds: Presence of biofilms? Wounds 2001, 13, 29–34. [Google Scholar]
- Nadell, C.D.; Xavier, J.B.; Levin, S.A.; Foster, K.R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008, 6, e14. [Google Scholar] [CrossRef]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; Eberl, L.; Tümmler, B. Quorum sensing: The power of cooperation in the world of Pseudomonas. Environ. Microbiol. 2005, 7, 459–471. [Google Scholar] [CrossRef]
- Hoang, T.T.; Sullivan, S.A.; Cusick, J.K.; Scihweizer, H.P. Beta-ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology 2002, 148, 3849–3856. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.; Karlyshev, A.V.; Fish, L.; Durant, E.L.; Winson, M.K.; Chhabra, S.R.; Williams, P.; Macintyre, S.; Stewart, G.S. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: Identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1997, 179, 5271–5281. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Winans, S.C. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 2001, 98, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Welch, M.; Todd, D.E.; Whitehead, N.A.; McGowan, S.J.; Bycroft, B.W.; Salmond, G.P. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 2000, 19, 631–641. [Google Scholar] [CrossRef]
- Hannan, S.; Ready, D.; Jasni, A.S.; Rogers, M.; Pratten, J.; Roberts, A.P. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Microbiol. 2010, 59, 345–349. [Google Scholar] [CrossRef]
- Klein, T.; Henn, C.; de Jong, J.C.; Zimmer, C.; Kirsch, B.; Maurer, C.K.; Pistorius, D.; Müller, R.; Steinbach, A.; Hartmann, R.W. Identification of small-molecule antagonists of the Pseudomonas aeruginosa transcriptional regulator PqsR: Biophysically guided hit discovery and optimization. ACS Chem. Biol. 2012, 7, 1496–1501. [Google Scholar] [CrossRef]
- Storz, M.P.; Maurer, C.K.; Zimmer, C.; Wagner, N.; Brengel, C.; de Jong, J.C.; Lucas, S.; Müsken, M.; Häussler, S.; Steinbach, A.; et al. Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J. Am. Chem. Soc. 2012, 134, 16143–16146. [Google Scholar] [CrossRef]
- Abraham, W. Going beyond the control of quorum-sensing to combat biofilm infections. Antibiotics 2016, 5, 3. [Google Scholar] [CrossRef]
- Scutera, S.; Zucca, M.; Savoia, D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin. Drug Discov. 2014, 9, 353–366. [Google Scholar] [CrossRef]
- Pearson, J.P.; Pesci, E.C.; Iglewski, B.H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef]
- Lee, K.; Yoon, S.S. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 2017, 27, 1053–1064. [Google Scholar] [CrossRef]
- Schuster, M.; Greenberg, E.P. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006, 296, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ni, N.; Li, M.; Wang, J.; Wang, B. Inhibitors and antagonists of bacterial quorum sensing. Med. Res. Rev. 2009, 29, 65–124. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Muraglia, E.; Bazzo, R.; Carfi, A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Urbanowski, M.L.; Greenberg, E.P. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 2004, 101, 15833–15839. [Google Scholar] [CrossRef]
- Gilbert, K.B.; Kim, T.H.; Gupta, R.; Greenberg, E.P.; Schuster, M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 2009, 73, 1072–1085. [Google Scholar] [CrossRef]
- Drees, S.L.; Fetzner, S. PqsE of Pseudomonas aeruginosa acts as pathway-specific thioesterase in the biosynthesis of alkylquinolone signaling molecules. Chem. Biol. 2015, 22, 611–618. [Google Scholar] [CrossRef]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A quorum sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [PubMed]
- Boursier, M.E.; Moore, J.D.; Heitman, K.M.; Shepardson-Fungairino, S.P.; Combs, J.B.; Koenig, L.C.; Shin, D.; Brown, E.C.; Nagarajan, R.; Blackwell, H.E. Structure-function analyses of the N-butanoyl l-homoserine lactone quorum sensing signal define features critical to activity in RhlR. ACS Chem. Biol. 2018, 13, 2655–2662. [Google Scholar] [CrossRef]
- Yang, L.; Nilsson, M.; Gjermansen, M.; Givskov, M.; Tolker-Nielsen, T. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 2009, 74, 1380–1392. [Google Scholar] [CrossRef]
- Pamp, S.J.; Tolker-Nielsen, T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2531–2539. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Senturk, S.; Ulusoy, S.; Bosgelmez-Tinaz, G.; Yagci, A. Quorum sensing and virulence of Pseudomonas aeruginosa during urinary tract infections. J. Infect. Dev. Ctries 2012, 6, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Déziel, E.; Gopalan, S.; Tampakaki, A.P.; Lépine, F.; Padfield, K.E.; Saucier, M.; Xiao, G.; Rahme, L.G. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: Multiple quorum-sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 2005, 55, 998–1014. [Google Scholar] [CrossRef] [PubMed]
- Lau, G.W.; Ran, H.; Kong, F.; Hassett, D.J.; Mavrodi, D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 2004, 72, 4275–4278. [Google Scholar] [CrossRef]
- Collier, D.N.; Anderson, L.; McKnight, S.L.; Noah, T.L.; Knowles, M.; Boucher, R.; Schwab, U.; Gilligan, P.; Pesci, E.C. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol. Lett. 2002, 215, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, L.A.; McKnight, S.L.; Kuznetsova, M.S.; Pesci, E.C.; Manoil, C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 6472–6480. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Dekimpe, V.; Déziel, E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Jensen, V.; Lons, D.; Zaoui, C.; Bredenbruch, F.; Meissner, A.; Dieterich, G.; Munch, R.; Haussler, S. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 2006, 188, 8601–8606. [Google Scholar] [CrossRef]
- Schafhauser, J.; Lepine, F.; McKay, G.; Ahlgren, H.G.; Khakimova, M.; Nguyen, D. The stringent response modulates 4-hydroxy-2-alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 1641–1650. [Google Scholar] [CrossRef]
- Oglesby, A.G.; Farrow, J.M., 3rd; Lee, J.; Tomaras, A.P.; Greenberg, E.P.; Pesci, E.C.; Vasil, M.L. The influence of iron on Pseudomonas aeruginosa physiology: A regulatory link between iron and quorum sensing. J. Biol. Chem. 2008, 283, 15558–15567. [Google Scholar] [CrossRef]
- Mattmann, M.E.; Shipway, P.M.; Heth, N.J.; Blackwell, H.E. Potent and selective synthetic modulators of a quorum sensing repressor in Pseudomonas aeruginosa identified from second-generation libraries of N-acylated L-homoserine lactones. Chem Biochem. 2011, 12, 942–949. [Google Scholar] [CrossRef]
- Patankar, A.V.; González, J.E. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 2009, 33, 739–756. [Google Scholar] [CrossRef]
- Parkins, M.D.; Ceri, H.; Storey, D.G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 2001, 40, 1215–1226. [Google Scholar] [CrossRef]
- Rodrigue, A.; Quentin, Y.; Lazdunski, A.; Méjean, V.; Foglino, M. Two-component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 2000, 8, 498–504. [Google Scholar] [CrossRef]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar] [CrossRef]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 2004, 28, 261–289. [Google Scholar] [CrossRef]
- Patriquin, G.M.; Banin, E.; Gilmour, C.; Tuchman, R.; Greenberg, E.P.; Poole, K. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 662–671. [Google Scholar] [CrossRef]
- Mattick, J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Dusane, D.H.; Zinjarde, S.S.; Venugopalan, V.P.; Mclean, R.J.C.; Weber, M.M.; Rahman, P.K.S.M. Quorum sensing: Implications on Rhamnolipid biosurfactant production. Biotechnol. Genet. Eng. Rev. 2010, 27, 159–184. [Google Scholar] [CrossRef]
- Boles, B.R.; Thoendel, M.; Singh, P.K. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 2005, 57, 1210–1223. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Stacey, R.E.; Dodd, C.; Cámara, M.; Williams, P.; Winzer, K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 2006, 8, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Tielker, D.; Hacker, S.; Loris, R.; Strathmann, M.; Wingender, J.; Wilhelm, S.; Rosenau, F.; Jaeger, K. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 2005, 151, 1313–1323. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Xu, K.D.; McFeters, G.A.; Stewart, P.S. Biofilm resistance to antimicrobial agents. Microbiology 2000, 146, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Cochran, W.L.; McFeters, G.A.; Stewart, P.S. Reduced susceptibility of thin Pseudomonas aeruginosa biofilms to hydrogen peroxide and monochloramine. J. Appl. Microbiol. 2000, 88, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.N.; Hutchison, J.B.; Melaugh, G.; Rodesney, C.; Roberts, A.E.L.; Irie, Y.; Jensen, P.Ø.; Diggle, S.P.; Allen, R.J.; Gordon, V.; et al. Role of multicellular aggregates in biofilm formation. mBio 2016, 7, e00237-16. [Google Scholar] [CrossRef]
- Sugimoto, S.; Okuda, K.; Miyakawa, R.; Sato, M.; Arita-Morioka, K.; Chiba, A.; Yamanaka, K.; Ogura, T.; Mizunoe, Y.; Sato, C. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci. Rep. 2016, 6, 25889. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Park, S.Y.; Park, H.J.; Moon, S.M.; Park, K.H.; Chong, Y.P.; Kim, M.N.; Kim, S.H.; Lee, S.O.; Kim, Y.S.; Woo, J.H.; et al. Impact of adequate empirical combination therapy on mortality from bacteremic Pseudomonas aeruginosa pneumonia. BMC Infect. Dis. 2012, 12, 308. [Google Scholar] [CrossRef]
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharm. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef]
- El Solh, A.A.; Alhajhusain, A. Update on the treatment of Pseudomonas aeruginosa pneumonia. J. Antimicrob. Chemother. 2009, 64, 229–238. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Speert, D.P. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and impact on treatment. Drug Resist. Updat. 2000, 3, 247–255. [Google Scholar] [CrossRef]
- Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Williams, T.C.; Oliver, J.D. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 2015, 23, 7–13. [Google Scholar] [CrossRef]
- Helaine, S.; Kugelberg, E. Bacterial persisters: Formation, eradication, and experimental systems. Trends Microbiol. 2014, 22, 417–424. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Kolar, M. Starvation- and antibiotics-induced formation of persister cells in Pseudomonas aeruginosa. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 2017, 161, 58–67. [Google Scholar] [CrossRef]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2011, 2, e00100-11. [Google Scholar] [CrossRef] [PubMed]
- Amato, S.M.; Fazen, C.H.; Henry, T.C.; Mok, W.W.K.; Orman, M.A.; Sandvik, E.L.; Volzing, K.G.; Brynildsen, M.P. The role of metabolism in bacterial persistence. Front. Microbiol. 2014, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Balamurugan, P.; Vasudevan, S.; Jadav, S.; Princy, S.A. Antimicrobial and antibiofilm potential of acyclic amines and diamines against multi-drug resistant Staphylococcus aureus. Front. Microbiol. 2017, 8, 1767. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilms: An emerging battleground in microbial communities. Anitimicrob. Resist. Infect. Control. 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Algburi, A.; Comito, N.; Kashtanov, D.; Dicks, L.M.T.; Chikindas, M.L. Control of biofilm formation: Antibiotics and Beyond. Appl. Environ. Microbiol. 2017, 83, e02508-16. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, E.V.T.; Lewenza, S.; Zenteno, S.R. Current research approaches to target biofilm infections. Postdoc. J. 2015, 3, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Hu, W.; Tian, Z.; Yuan, D.; Yi, G.; Zhou, Y.; Cheng, Q.; Zhu, J.; Li, M. Developing natural products as potential anti-biofilm agents. Chin. Med. 2019, 14, 11. [Google Scholar] [CrossRef]
- Al-Haidari, R.A.; Shaaban, M.I.; Ibrahim, S.R.M.; Mohamed, G.A. Anti-quorum sensing activity of some medicinal plants. Afr. J. Tradit. Complement Altern. Med. 2016, 13, 67–71. [Google Scholar] [PubMed]
- Musthafa, K.S.; Ravi, A.V.; Annapoorani, A.; Packiavathy, I.S.V.; Pandian, S.K. Evaluation of anti-quorum-sensing activity of edible plants and fruits through inhibition of the N-acyl-homoserine lactone system in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 2010, 56, 333–339. [Google Scholar] [CrossRef]
- Vasavi, H.S.; Arun, A.B.; Rekha, P.D. Anti-quorum sensing activity of flavonoid rich fraction from Centella asiatica L. against Pseudomonas aeruginosa PAO1. J. Microbiol. Immunol. Infect. 2016, 49, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabi, N.A.; Balachandran, C.; Raj, M.K.; Duraipandiyan, V.; Muthukumar, C.; Ignacimuthu, S.; Khan, I.A.; Rajput, V.S. Antimicrobial, antimycobacterial and antibiofilm properties of Couroupita guianensis Aubl. fruit extract. BMC Complement. Altern. Med. 2012, 12, 242. [Google Scholar] [CrossRef]
- Perumal, S.; Mahmud, R. Chemical analysis, inhibition of biofilm formation and biofilm eradication potential of Euphorbia hirta L. against clinical isolates and standard strains. BMC Complement. Altern. Med. 2013, 13, 346. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Rasmussen, T.B.; Christophersen, L.; Calum, H.; Hentzer, M.; Hougen, H.P.; Rygaard, J.; Moser, C.; Eberl, L.; et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 2005, 151, 3873–3880. [Google Scholar] [CrossRef]
- Zahin, M.; Hasan, S.; Aqil, K.; Khan, M.S.A.; Husain, F.M.; Ahmad, I. Screening of certain medicinal plants from India for their anti-quorum sensing activity. Indian J. Exp. Biol. 2010, 48, 1219–1224. [Google Scholar]
- Song, Z.; Kong, K.F.; Wu, H.; Maricic, N.; Ramalingam, B.; Priestap, H.; Schneper, L.; Quirke, J.M.E.; Høiby, N.; Mathee, K. Panax ginseng has anti-infective activity against opportunistic pathogen Pseudomonas aeruginosa by inhibiting quorum sensing, a bacterial communication process critical for establishing infection. Phytomedicine 2010, 17, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.S.P.; Brandão, D.O.; Chaves, T.P.; Filho, A.L.N.F.; de B Costa, E.M.M.; Santos, V.L.; Medeiros, A.C.D. Study bioprospecting of medicinal plant extracts of the semiarid northeast: Contribution to the control of oral microorganisms. Evid. Based Complement. Alternat. Med. 2012, 2, 681207. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, T.; Yin, W.F.; Chan, K.G. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa PA01 by ayurveda spice clove (Syzygium aromaticum) bud extract. Sensors 2012, 12, 4016–4030. [Google Scholar] [CrossRef]
- Taganna, J.C.; Quanico, J.P.; Perono, R.M.G.; Amor, E.C.; Rivera, W. Tannin-rich fraction from Terminalia catappa inhibits quorum sensing (QS) in Chromobacterium violaceum and the QS-controlled biofilms maturation and LasA staphylolytic activity in Pseudomonas aeruginosa. J. Ethnopharmacol. 2011, 134, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Juarez, I.C.; Contreras, R.G.; Guadarrama, N.V.; Hernandez, M.S.; Vazquez, M.M. Amphypterygium adstringens anacardic acid mixture inhibits quorum sensing controlled virulence factors of Chromobacterium violaceum and Pseudomonas aeruginosa. Arch. Med. Res. 2013, 44, 488–494. [Google Scholar] [CrossRef]
- Sarkar, R.; Chaudhary, S.K.; Sharma, A.; Yadav, K.; Nema, N.; Sekhoacha, M.; Karmakar, S.; Braga, F.C.; Matsabisa, M.G.; Mukherjee, P.K.; et al. Anti-biofilm activity of Marula- a study with the standardized bark extract. J. Ethnopharmacol. 2014, 154, 170–175. [Google Scholar] [CrossRef]
- Vattem, D.A.; Mihalik, K.; Crixell, S.H.; McLean, R.J.C. Dietary phytochemicals as quorum sensing inhibitors. J. Fitoterapia 2007, 78, 302–310. [Google Scholar] [CrossRef]
- Chong, Y.M.; Yin, W.F.; Ho, C.Y.; Mustafa, M.R.; Hamid, A.; Hadi, A.; Awang, K.; Narrima, P.; Koh, C.L.; Appleton, D.R.; et al. Malabaricone C from Myristica cinnamomea exhibits anti-quorum sensing activity. J. Nat. Prod. 2011, 74, 2261–2264. [Google Scholar] [CrossRef]
- Tan, L.Y.; Yin, W.F.; Chan, K.G. Silencing quorum sensing through extracts of Melicope lunu-ankenda. Sensors 2012, 12, 4339–4351. [Google Scholar] [CrossRef]
- Priya, K.; Yin, W.F.; Chan, K.G. Anti-quorum sensing activity of the traditional Chinese herb, Phyllanthus amarus. Sensors 2013, 13, 14558–14569. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.V.P.I.; Palani, A.; Ramaswamy, B.R.; Shunmugiah, K.P.; Arumugam, V.R. Antiquorum sensing and antibiofilm potential of Capparis spinosa. J. Arch. Med. Res. 2011, 42, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Lee, S.J.; Park, J.Y.; Reza, M.A.; Kim, T.H.; Lee, K.J.; Suh, J.W.; Park, S.C. Modulation of quorum sensing-controlled virulence factors by Nymphaea tetragona (waterlily) extract. J. Ethnopharmcol. 2015, 174, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Bhathena, Z. Broad spectrum anti-quorum sensing activity of tannin-rich crude extracts of Indian medicinal plants. Scientifica 2016, 5, 5823013. [Google Scholar] [CrossRef]
- Mombeshora, M.; Chi, G.F.; Mukanganyama, S. Antibiofilm activity of extract and a compound isolated from Triumfetta welwitschii against Pseudomonas aeruginosa. Biochem. Res. Int. 2021, 14, 9946183. [Google Scholar]
- Al-Youef, H.M.; Alam, P.; Khanam, Z.; Amnia, M.; Hassan, W.H.B. Corchorus olitorius aqueous extract attenuates quorum-sensing regulated virulence factor production and biofilm formation. Asian Pac. J. Trop. Biomed. 2021, 11, 66–73. [Google Scholar] [CrossRef]
- Jalli, N.; Sri, K.V.S.; Hnamte, S.; Pattnaik, S.; Paramanantham, P.; Siddhardha, B. Antioxidant, anti-quorum sensing and anti-biofilm potential of ethanolic leaf extract of Phrylium capitatum and Dryptes indica. Asian Pac. J. Biomed. 2019, 9, 323–332. [Google Scholar]
- Li, S.; Yao, J.; Li, H. Plantain Herb Extracts significantly attenuate the quorum sensing-controlled virulence factors and inhibit biofilm formation in Pseudomonas aeruginosa PAO1. E3S Web Conf. 2019, 78, 01004. [Google Scholar] [CrossRef]
- Topa, S.H.; Palombo, E.A.; Kingshott, P.; Blackall, L.L. Activity of Cinnamaldehyde on quorum sensing and biofilm susceptibility to antibiotics in Pseudomonas aeruginosa. Microorganisms 2020, 8, 455. [Google Scholar] [CrossRef]
- Hnamte, S.; Subhaswaraj, P.; Ranganathan, S.K.; Ampasala, D.R.; Muralitharan, G.; Siddhardha, B. Antiquorum sensing and antibiofilm potential of Anogeissus acuminata and Mallotus roxburghianus Muell. against Pseudomonas aeruginosa PAO1. J. Microbiol. Biotechnol. Food Sci. 2019, 8, 1135–1140. [Google Scholar]
- Jalli, N.; Sri, K.V.S.; Hnamte, S.; Pattnaik, S.; Paramanantham, P.; Siddhardha, B. Experimental investigations on Camellia kissi wall. for antioxidant, anti-quorum sensing and anti-biofilm activities. J. Microbiol. Biotechnol. Food Sci. 2020, 9, 736–741. [Google Scholar] [CrossRef]
- Hnamte, S.; Subhaswaraj, P.; Ranganathan, S.K.; Ampasala, D.R.; Muralitharan, G.; Siddhardha, B. Methanolic extract of Plectranthus tenuiflorus attenuates quorum sensing mediated virulence and biofilm formation in Pseudomonas aeruginosa PAO1. J. Pure Appl. Microbiol. 2018, 12, 1985–1996. [Google Scholar] [CrossRef]
- Jovanović, M.; Morić, I.; Nikolić, B.; Pavić, A.; Svirčev, E.; Šenerović, L.; Mitić-Ćulafić, D. Anti-virulence potential and in vivo toxicity of Persicaria maculosa and Bistorta officinalis extracts. Molecules 2020, 25, 1811. [Google Scholar] [CrossRef]
- Famuyide, I.M.; Aro, A.O.; Fasina, F.O.; Eloff, J.N.; McGaw, L.J. Antibacterial and antibiofilm activity of acetone leaf extracts of nine under-investigated south African Eugenia and Syzygium (Myrtaceae) species and their selectivity indices. BMC Complement Altern. Med. 2019, 19, 141. [Google Scholar] [CrossRef] [PubMed]
- Alam, K.; Farraj, D.A.A.; Mah-e-Fatima, S.; Yameen, M.A.; Elshikh, M.S.; Alkufeidy, R.M.; Naqvi, T.A. Anti-biofilm activity of plant derived extracts against infectious pathogen-Pseudomonas aeruginosa PAO1. J. Infect. Public Health 2020, 13, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Emam, M.; Abdel-Haleem, D.R.; Salem, M.M.; Abdel-Hafez, L.J.M.; Latif, R.R.A.; Farag, S.M.; El Raey, M.A. Phytochemical profiling of Lavandula coronopifolia Poir. aerial parts extract and its larvicidal, antibacterial, and antibiofilm activity against Pseudomonas aeruginosa. Molecules 2021, 26, 1710. [Google Scholar] [PubMed]
- Jahan, M.; Abuhena, M.D.; Azad, A.K.; Karim, M.M. In vitro antibacterial and antibiofilm activity of selected medicinal plants and spices extracts against multidrug resistant Pseudomonas aeruginosa. J. Pharmacogn. Phytochem. 2018, 7, 2114–2121. [Google Scholar]
- Torres, C.E.; Lenon, G.; Craperi, D.; Wilting, R.; Blanco, A. Enzymatic treatment for preventing biofilm formation in the paper industry. Appl. Microbiol. Biotechnol. 2011, 92, 95–103. [Google Scholar] [CrossRef]
- Kovach, K.N.; Fleming, D.; Wells, M.J.; Rumbaugh, K.P.; Gordon, V.D. Specific disruption of established P. aeruginosa biofilms using polymer-attacking enzymes. Langmuir 2020, 36, 1585–1595. [Google Scholar] [CrossRef]
- Dong, Y.H.; Xu, J.L.; Li, X.Z.; Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar] [CrossRef]
- Bräuer, N.W.; Kisch, M.J.; Pinnow, N.; Liese, A.; Schmitz, R.A. Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 2016, 7, 1098. [Google Scholar]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2015, 201, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Gawande, P.V.; Clinton, A.P.; Lovetri, K.; Yakandawala, N.; Rumbaugh, K.P.; Madhyastha, S. Antibiofilm efficacy of DispersinB® wound spray used in combination with a silver wound dressing. Microbiol. Insights 2014, 7, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Gawande, P.V.; Leung, K.P.; Madhyastha, S. Antibiofilm and antimicrobial efficacy of DispersinB®-KSL-W peptide-based wound gel against chronic wound infection associated bacteria. Curr. Microbiol. 2014, 68, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Demidova-Rice, T.N.; Hamblin, M.R.; Herman, I.M. Acute and impaired wound healing: Pathophysiology and current methods for drug delivery, part 1: Normal and chronic wounds: Biology, causes, and approaches to care. Adv. Skin Wound Care 2012, 25, 304–314. [Google Scholar] [CrossRef]
- Ramundo, J.; Gray, M. Enzymatic wound debridement. J. Wound Ostomy Cont. Nurs. 2008, 35, 273–280. [Google Scholar] [CrossRef]
- Falabella, A.F. Debridement and wound bed preparation. Dermatol. Ther. 2006, 19, 317–325. [Google Scholar] [CrossRef]
- Ge, L.; Zhao, Y.S.; Mo, T.; Li, J.R.; Li, P. Immobilization of glucose oxidase in electrospun nanofibrous membranes for food preservation. Food Control. 2012, 26, 188–193. [Google Scholar] [CrossRef]
- Hansen, E.H.; Albertsen, L.; Schafer, T.; Johansen, C.; Frisvad, J.C.; Molin, S.; Gram, L. Curvularia haloperoxidase: Antimicrobial activity and potential application as a surface disinfectant. Appl. Environ. Microbiol. 2003, 69, 4611–4617. [Google Scholar] [CrossRef][Green Version]
- Barton, N.; Robertson, D.; Chang, K.; Elkins, J. Enzymes and the Nucleic Acids Encoding Them and Methods for Making and Using Them. WIPO Patent WO2004066945, 8 December 2004. [Google Scholar]
- Fagerlund, A.; Langsrud, S.; Heir, E.; Mikkelsen, M.I.; Møretrø, T. Biofilm matrix composition affects the susceptibility of food associated staphylococci to cleaning and disinfection agents. Front. Microbiol. 2016, 7, 856. [Google Scholar] [CrossRef]
- Kalpana, B.J.; Aarthy, S.; Pandian, S.K. Antibiofilm activity of α-amylase from Bacillus subtilis S8-18 against biofilm forming human bacterial pathogens. Appl. Biochem. Biotechnol. 2012, 167, 1778–1794. [Google Scholar] [CrossRef]
- Lamppa, J.W.; Griswold, K.E. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob. Agents Chemother. 2013, 57, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Grover, N.; Plaks, J.G.; Summers, S.R.; Chado, G.R.; Schurr, M.J.; Kaar, J.L. Acylase-containing polyurethane coatings with anti-biofilm activity. Biotechnol. Bioeng. 2016, 113, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, K.; Fernandes, M.M.; Mendoza, E.; Tzanov, T. Enzyme multilayer coatings inhibit Pseudomonas aeruginosa biofilm formation on urinary catheters. Appl. Microbiol. Biotechnol. 2015, 99, 4373–4385. [Google Scholar] [CrossRef]
- Vogel, J.; Havinga, M.W.; Setroikromo, R.; Quax, W.J. Immobilized acylase PvdQ reduces Pseudomonas aeruginosa biofilm formation on PDMS silicone. Front. Chem. 2020, 8, 1–9. [Google Scholar] [CrossRef]
- Hukić, M.; Seljmo, D.; Ramovic, A.; Ibrišimović, M.A.; Dogan, S.; Hukic, J.; Bojic, E.F. The effect of lysozyme on reducing biofilms by Staphylococcus aureus, Pseudomonas aeruginosa, and Gardnerella vaginalis: An in vitro examination. Microb. Drug Resist. 2018, 24, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Sebaa, S.; Hizette, N.; Otmani, Z.B.; Courtois, P. Dose-dependent effect of lysozyme upon Candida albican biofilm. Mol. Med. Rep. 2017, 15, 1135–1142. [Google Scholar] [CrossRef]
- Shukla, S.K.; Rao, T.S. Staphylococcus aureus biofilm removal by targeting biofilm-associated extracellular proteins. Indian J. Med. Res. 2017, 146, S1–S8. [Google Scholar]
- Eladawy, M.; Mowafy, M.E.; Sokkary, M.E.; Barwa, R. Effects of lysozyme, proteinase K, and cephalosporins on biofilm formation by clinical isolates of Pseudomonas aeruginosa. Interdiscip. Perspect. Infect. Dis. 2020, 4, 6156720. [Google Scholar] [CrossRef]
- Bijtenhoorn, P.; Mayerhofer, H.; Dieckmann, J.M.; Utpatel, C.; Schipper, C.; Hornung, C.; Szesny, M.; Grond, S.; Thürmer, A.; Brzuszkiewicz, E.; et al. A novel metagenomic Short-Chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE 2011, 6, e26278. [Google Scholar] [CrossRef] [PubMed]
- Witzgall, F.; Depke, T.; Hoffmann, M.; Empting, M.; Brönstrup, M.; Müller, R.; Blankenfeldt, W. The alkylquinolone repertoire of Pseudomonas aeruginosa is linked to structural flexibility of the FabH-like 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) biosynthesis enzyme PqsBC. Chembiochem 2018, 19, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, J.T.; Galloway, W.R.; Welch, M.; Spring, D.R. Microwave-assisted preparation of the quorum sensing molecule 2-heptyl-3-hydroxy-4(1H)-quinolone and structurally related analogs. Nat. Protoc. 2012, 7, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Banar, M.; Emaneini, M.; Beigverdi, R.; Pirlar, R.F.; Farahani, N.N.; van Leeuwen, W.B.; Abalameli, F. The efficacy of lyticase and β-glucosidase enzymes on biofilm degradation of Pseudomonas aeruginosa strains with different gene profiles. BMC Microbiol. 2019, 19, 291. [Google Scholar] [CrossRef] [PubMed]
- Daboor, S.M.; Raudonis, R.; Cohen, A.; Rohde, J.R.; Cheng, Z. Marine bacteria, a source for alginolytic enzyme to disrupt Pseudomonas aeruginosa biofilms. Mar. Drugs 2019, 17, 307. [Google Scholar] [CrossRef] [PubMed]
- Mion, S.; Rémy, B.; Plener, L.; Brégeon, F.; Chabrière, E.; Daudé, D. Quorum quenching lactonase strengthens bacteriophage and antibiotic arsenal against Pseudomonas aeruginosa clinical isolates. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Snarr, B.D.; Baker, P.; Bamford, N.C.; Sato, Y.; Liu, H.; Lehoux, M.; Gravelat, F.N.; Ostapska, H.; Baistrocchi, S.R.; Cerone, R.P.; et al. Microbial glycoside hydrolases as antibiofilm agents with cross-kingdom activity. Proc. Natl. Acad. Sci. USA 2017, 114, 7124–7129. [Google Scholar] [CrossRef]
- Stiefel, P.; Mauerhofer, S.; Schneider, J.; Weber, K.M.; Rosenberg, U.; Ren, Q. Enzymes enhance biofilm removal efficiency of cleaners. Antimicrob. Agents Chemother. 2016, 60, 3646–3652. [Google Scholar] [CrossRef]
- Banar, M.; Emaneini, M.; Satarzadeh, M.; Abdellahi, N.; Beigverdi, R.; van Leeuwen, W.B.; Jabalameli, F. Evaluation of mannosidase and trypsin enzymes effects on biofilm production of Pseudomonas aeruginosa isolated from burn wound infections. PLoS ONE 2016, 11, e0164622. [Google Scholar] [CrossRef] [PubMed]
- Hymes, S.R.; Randis, T.M.; Sun, T.Y.; Ratner, A.J. DNase inhibits Gardnerella vaginalis biofilms in vitro and in vivo. J. Infect. Dis. 2013, 207, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209. [Google Scholar] [CrossRef]
- Parks, Q.M.; Young, R.L.; Poch, K.R.; Malcolm, K.C.; Vasil, M.L.; Nick, J.A. Neutrophil enhancement of Pseudomonas aeruginosa biofilm development: Human F-actin and DNA as targets for therapy. J. Med. Microbiol. 2009, 58, 492–502. [Google Scholar] [CrossRef]
- Darouiche, R.O.; Mansouri, M.D.; Gawande, P.V.; Madhyastha, S. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB® combination. J. Antimicrob. Chemother. 2009, 64, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, A.S.; Nelis, H.; Coenye, T. Investigating the role of matrix components in protection of Burkholderia cepacia complex biofilms against tobramycin. J. Cyst. Fibros. 2014, 13, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, J.H.; Cho, M.H.; Herzberg, M.; Lee, J. Acceleration of protease effect on Staphylococcus aureus biofilm dispersal. FEMS Microbiol. Lett. 2012, 335, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Nijland, R.; Hall, M.J.; Burgess, J.G. Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS ONE 2010, 5, e15668. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.; Hill, P.J.; Snarr, B.D.; Alnabelseya, N.; Pestrak, M.J.; Lee, M.J.; Jennings, L.K.; Tam, J.; Melnyk, R.A.; Parsek, M.R.; et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016, 2, e1501632. [Google Scholar] [CrossRef] [PubMed]
- Kiran, S.; Sharma, P.; Harjai, K.; Capalash, N. Enzymatic quorum quenching increases antibiotic susceptibility of multidrug resistant Pseudomonas aeruginosa. Iran. J. Microbiol. 2011, 3, 1–12. [Google Scholar] [PubMed]
- Tielen, P.; Rosenau, F.; Wilhelm, S.; Jaeger, K.E.; Flemming, H.C.; Wingender, J. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas aeruginosa. Microbiology 2010, 156, 2239–2252. [Google Scholar] [CrossRef]
- Kaplan, J.B. Therapeutic potential of biofilm-dispersing enzymes. Int. J. Artif. Organs 2009, 32, 545–554. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, L.; Wu, H.; Zhao, C.; Gong, Q.; Yu, W. Cladodionen is a potential quorum sensing inhibitor against Pseudomonas aeruginosa. Mar. Drugs 2020, 18, 205. [Google Scholar] [CrossRef]
- Sadiq, S.; Rana, N.F.; Zahid, M.A.; Zargaham, M.K.; Tanweer, T.; Batool, A.; Naeem, A.; Nawaz, A.; Rizwan-ur-Rehman; Muneer, Z.; et al. Virtual screening of FDA-approved drugs against LasR of Pseudomonas aeruginosa for antibiofilm potential. Molecules 2020, 25, 3723. [Google Scholar] [CrossRef] [PubMed]
- Baldelli, V.; D’Angelo, F.; Pavoncello, V.; Fiscarelli, E.V.; Visca, P.; Rampioni, G.; Leoni, L. Identification of FDA-approved antivirulence drugs targeting the Pseudomonas aeruginosa quorum sensing effector protein PqsE. Virulence 2020, 11, 652–668. [Google Scholar] [CrossRef]
- Abelyan, N.; Grabski, H.; Tiratsuyan, S. In silico screening of flavones and its derivatives as potential inhibitors of quorum-sensing regulator LasR of Pseudomonas aeruginosa. J. Mol. Biol. 2020, 54, 153–163. [Google Scholar] [CrossRef]
- Mellini, M.; Muzio, E.D.; D’Angelo, F.; Baldelli, V.; Ferrillo, S.; Visca, P.; Leoni, L.; Polticelli, F.; Rampioni, G. In silico selection and experimental validation of FDA approved drugs as anti-quorum sensing agents. Front. Microbiol. 2019, 10, 2355. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.D.; Kharkar, P.S.; Sahu, N.U.; Peerzada, Z.; Desai, K.B. Potassium 2-methoxy-4-vinylphenolate: A novel hit exhibiting quorum-sensing inhibition in Pseudomonas aeruginosa via LasIR/RhlIR circuitry. RSC Adv. 2019, 9, 40228–40239. [Google Scholar] [CrossRef]
- Nain, Z.; Sayed, S.B.; Karim, M.M.; Islam, M.A.; Adhikari, U.K. Energy-optimized pharmacophore coupled virtual screening in the discovery of quorum sensing inhibitors of LasR protein of Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. 2019, 38, 5374–5388. [Google Scholar] [CrossRef]
- Singh, S.; Bhatia, S. In silico identification of albendazole as a quorum sensing inhibitor and its in vitro verification using CviR and LasB receptors based assay systems. BioImpacts 2018, 8, 201–209. [Google Scholar] [CrossRef]
- Paczkowski, J.E.; Mukherjee, S.; McCready, A.R.; Cong, J.P.; Aquino, C.J.; Kim, H.; Henke, B.R.; Smith, C.D.; Bassler, B.L. Flavonoids supress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J. Biol. Chem. 2017, 292, 4064–4076. [Google Scholar] [CrossRef]
- Xu, Y.; Tong, X.; Sun, P.; Bi, L.; Lin, K. Virtual screening and biological evaluation of biofilm inhibitors on dual targets in quorum sensing system. Future Med. Chem. 2017, 9, 1983–1994. [Google Scholar] [CrossRef]
- Jha, S.K.; Rashmi, S.; Shubhra, R.S.; Singh, H.R. High throughput screening of quorum sensing inhibitors based lead molecules for Pseudomonas aeruginosa associated infections. Int. J. Pharm. Clin. Res. 2014, 6, 214–220. [Google Scholar]
- Tan, S.Y.; Chua, S.L.; Chen, Y.; Rice, S.A.; Kjelleberg, S.; Nielsen, T.E.; Yang, L.; Givskova, M. Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob. Agents Chemother. 2013, 57, 5629–5641. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Lo, C.; Walsh, C.; Hiller, L.; Marculescu, R. In silico evaluation of the impacts of quorum sensing inhibition (QSI) on strain competition and development of QSI resistance. Sci. Rep. 2016, 6, 35136. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, P.; Kumar, R.; Bhardwaj, A. dPABBs: A novel in silico approach for predicting and designing anti-biofilm peptides. Sci. Rep. 2016, 6, 21839. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, S.H.; Byun, Y.; Park, H.D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 2015, 5, 8656. [Google Scholar] [CrossRef]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin influences quorum sensing in food borne bacteria: In vitro and in silico evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef]
- Xu, Z.; Fang, X.; Wood, T.K.; Huang, Z.J. A systems-level approach for investigating Pseudomonas aeruginosa biofilm formation. PLoS ONE 2013, 8, e57050. [Google Scholar]
- Sahner, J.H.; Brengel, C.; Storz, M.P.; Groh, M.; Plaza, A.; Müller, R.; Hartmann, R.W. Combining in Silico and biophysical methods for the development of Pseudomonas aeruginosa quorum sensing inhibitors: An alternative approach for structure-based drug design. J. Med. Chem. 2013, 56, 8656–8664. [Google Scholar] [CrossRef]
- Amin, E.A.; Welsh, W.J. A preliminary in silico lead series of 2-phthalimidinoglutaric acid analogues designed as MMP-3 inhibitors. J. Chem. Inf. Model. 2006, 46, 2104–2109. [Google Scholar] [CrossRef]
- Ekins, S.; Mestres, J.; Testa, B. In silico pharmacology for drug discovery:applications to tagrets and beyond. Br. J. Pharmacol. 2007, 152, 21–37. [Google Scholar] [CrossRef]

| Polysaccharide | Gene | Function | References |
|---|---|---|---|
| Alginate | AlgD | Sugar nucleotide production | [71] |
| Alg8 | Subunit polymerization | [72] | |
| Alg44 | c-di-GMP binding | [73] | |
| AlgK | Outer membrane protein/secretion | [74] | |
| AlgE | Outer membrane protein/secretion | [74] | |
| AlgG | Epimerase/modification | [75] | |
| AlgX | Epimerase/modification | [76] | |
| AlgL | Hydrolase/lyase | [77] | |
| AlgI | O-Acetylation | [78] | |
| AlgJ | O-Acetylation | [76] | |
| AlgF | O-Acetylation | [78] | |
| AlgA | Sugar nucleotide production | [79] | |
| Pel | PelA | Hydrolase/lyase | [80] |
| PelB | Outer membrane protein/secretion | [81] | |
| PelC | Outer membrane protein/secretion | [82] | |
| PelD | c-di-GMP binding | [83] | |
| PelE | Subunit polymerization | [84] | |
| PelF | Glycosyl transferase | [85] | |
| PelG | Inner membrane protein | [86] | |
| Psl | PslA | Subunit polymerization | [87] |
| PslB | Sugar nucleotide production | [79] | |
| PslC | Glycosyl transferase | [72] | |
| PslD | Outer membrane protein/secretion | [88] | |
| PslE | Wzz/Wzc like protein | [89] | |
| PslF | Glycosyl transferase | [90] | |
| PslG | Hydrolase/lyase | [91] | |
| PslH | Glycosyl transferase | [90] | |
| PslI | Glycosyl transferase | [90] | |
| PslJ | Inner membrane protein | [86] | |
| PslK | Inner membrane protein | [86] | |
| PslL | Inner membrane protein | [86] |
| S. No | Plant Species | Plant Part | Extract | References |
|---|---|---|---|---|
| 1 | Allium cepa | Outer scales | Methanol | [168] |
| 2 | Allium sativa | Bulbs | Methanol | [168] |
| 3 | Ananas comosus | Fruit | Aqueous | [169] |
| 4 | Centella asiatica | Leaves | Ethanol | [170] |
| 5 | Citrus sinensis | Seeds | Methanol | [168] |
| 6 | Coriandrum sativum | Fruit | Methanol | [168] |
| 7 | Couroupita guianensis | Fruit | Chloroform | [171] |
| 8 | Elettaria cardamomum | Seeds | Methanol | [168] |
| 9 | Euphorbia hirta L. | Aerial parts | Methanol | [172] |
| 10 | Garlic | Bulbs | Toluene | [173] |
| 11 | Hemidesmus indicus (L.) | Root | Ethanol | [174] |
| 12 | Holarrhena antidysenterica | Bark | Ethanol | [174] |
| 13 | Laurus nobilis | Leaves | Methanol | [168] |
| 14 | Mangifera indica L. | Seed | Ethanol | [174] |
| 15 | Manilkara zapota | Fruit | Aqueous | [169] |
| 16 | Mentha longifolia | Aerial part | Methanol | [168] |
| 17 | Musa paradiciaca | Stem | Aqueous | [169] |
| 18 | Ocimum sanctum | Leaves | Aqueous | [169] |
| 19 | Panax notoginseng | Roots | Aqueous | [175] |
| 20 | Psidium guajava | Leaves | Methanol | [168] |
| 21 | Psoralea corylifolia L. | Seeds | Ethanol | [174] |
| 22 | Senecio brasiliensis | Stem bark | Ethanol/Aqueous | [176] |
| 23 | Syzygium aromaticum | Bud | Hexane, Chloroform, Methanol | [177] |
| 24 | Terminalia catappa | Leaves | Methanol | [178] |
| 25 | Amphypterygium adstringens | Stem bark | Hexane | [179] |
| 26 | Sclerocarya birrea | Stem bark | Methanol | [180] |
| 27 | Ocimum basilica | Whole plant | Aqueous | [181] |
| 28 | Brassica oleracea | Whole plant | Aqueous | [181] |
| 29 | Zingiber officinale | Whole plant | Aqueous | [181] |
| 30 | Myristica cinnamomea | Bark | Methanol | [182] |
| 31 | Melicope lunu-ankenda | Leaves | Hexane, Chloroform, Methanol | [183] |
| 32 | Psidium guajava | Leaves | Methanol | [168] |
| 33 | Phyllanthus amarus | Whole plant | Hexane, Chloroform, Methanol | [184] |
| 34 | Capparis spinosa | Dried fruit | Methanol | [185] |
| 35 | Thymus sp. | Whole plant | Aqueous | [181] |
| 36 | Nymphaea tetragona | Whole plant | Aqueous | [186] |
| 37 | Terminalia bellirica | Fruits | Methanol | [187] |
| 38 | Terminalia chebula | Fruits | Methanol | [187] |
| 39 | Syzygium cumini | Seeds | Methanol | [187] |
| 40 | Sclerocarya birrea | Bark | Methanol | [180] |
| 41 | Punica granatum L. | Pericarp | Ethanol | [174] |
| 42 | Triumfetta welwitschii | leaves | Dichloromethane: methanol | [188] |
| 43 | Corchorus olitorius | stem | Ethanol | [189] |
| 44 | Phrynium capitatum | Leaves | Ethanol | [190] |
| 45 | Dryptes indica | Leaves | Ethanol | [190] |
| 46 | Plantain herb | Whole plant | Ethanol | [191] |
| 47 | Cinnamomum camphora | Bark | Distilled water | [192] |
| 48 | Centella asiatica | Leaves | Ethanol | [170] |
| 49 | Anogeissus acuminata | Whole plant | Methanol | [193] |
| 50 | Mallotus roxburghianus Muell | Whole plant | Ethanol | [193] |
| 51 | Camellia kissi wall. | Leaves | Methanol | [194] |
| 52 | Plectranthus tenuiflorus | Leaves | Methanol | [195] |
| 53 | Persicaria maculosa | Aerial parts | Ethanol | [196] |
| 54 | Bistorta officinalis | Rhizome | Ethanol | [196] |
| 55 | Syzygium legatii | Leaves | Acetone | [197] |
| 56 | Syzygium masukuense | Leaves | Acetone | [197] |
| 57 | Syzygium species A | Leaves | Acetone | [197] |
| 58 | Berginia ciliate | rhizome with skin | Methanol | [198] |
| 59 | Lavandulacoronopifolia | aerial parts | Methanol: water | [199] |
| 60 | Centella asiatica | Leaves | Methanol | [200] |
| 61 | Mentha spicata | Leaves | Methanol | [200] |
| 62 | Azadirachta indica | Leaves | Methanol | [200] |
| 63 | Psidium guajava | Leaves | Methanol | [200] |
| 64 | Syzygium aromaticum | Whole part | Ethyl acetate | [200] |
| 65 | Cinnamomum zeylanicum | Whole part | Ethyl acetate | [200] |
| Class of Enzyme | Example | Target | References |
|---|---|---|---|
| Oxidoreductases | Glucose oxidase, Curvularia haloperoxidase | Directly or indirectly retarding bacterial growth by production of H2O2 | [211,212] |
| Transferases | Transaminase | EPS matrix | [213] |
| Hydrolases | AiiA, α-amylase, Proteinase K | QS molecules, Exopolysaccharides, Exoproteins | [203,214,215] |
| Lyases | Alginate lyase | Exopolysaccharides | [216] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vetrivel, A.; Ramasamy, M.; Vetrivel, P.; Natchimuthu, S.; Arunachalam, S.; Kim, G.-S.; Murugesan, R. Pseudomonas aeruginosa Biofilm Formation and Its Control. Biologics 2021, 1, 312-336. https://doi.org/10.3390/biologics1030019
Vetrivel A, Ramasamy M, Vetrivel P, Natchimuthu S, Arunachalam S, Kim G-S, Murugesan R. Pseudomonas aeruginosa Biofilm Formation and Its Control. Biologics. 2021; 1(3):312-336. https://doi.org/10.3390/biologics1030019
Chicago/Turabian StyleVetrivel, Aishwarya, Monica Ramasamy, Preethi Vetrivel, Santhi Natchimuthu, Shobana Arunachalam, Gon-Sup Kim, and Rajeswari Murugesan. 2021. "Pseudomonas aeruginosa Biofilm Formation and Its Control" Biologics 1, no. 3: 312-336. https://doi.org/10.3390/biologics1030019
APA StyleVetrivel, A., Ramasamy, M., Vetrivel, P., Natchimuthu, S., Arunachalam, S., Kim, G.-S., & Murugesan, R. (2021). Pseudomonas aeruginosa Biofilm Formation and Its Control. Biologics, 1(3), 312-336. https://doi.org/10.3390/biologics1030019







