Abstract
Background/Objectives: Bacteriophages are considered promising alternatives for the treatment of multidrug-resistant (MDR) Pseudomonas aeruginosa infections. Methods: Five bacteriophages with lytic activity against MDR P. aeruginosa were isolated from lake and sewage samples and characterized for their biological properties, host range, and efficacy in biofilm and in vitro infection models. Results: The phages displayed broad host ranges, producing zones of lysis in 40–53% of MDR isolates. The average burst size was 112 ± 70 PFU per cell. All phages, either individually or in combination, inhibited biofilm formation and were capable of disrupting preformed biofilms. While treatment with single phages led to bacterial regrowth, the cocktail of all five phages achieved complete bacterial lysis with no regrowth observed. In an in vitro wound and burn infection model, the phage cocktail significantly enhanced cell proliferation and promoted healing. Transmission electron microscopy (TEM) analysis identified phage PA2 as a Myovirus based on its morphology. Conclusions: The phage isolates demonstrated strong activity in multiple in vitro models, effectively targeting both planktonic and biofilm-associated P. aeruginosa. Notably, the five-phage combination prevented the emergence of bacterial resistance, supporting its potential as a biocontrol strategy against MDR P. aeruginosa.
1. Introduction
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen with a relatively large genome (5.5–7 Mbp) [1], which allows it to adapt and thrive in diverse ecological niches. It produces a wide range of virulence factors [2], including biofilm-forming ability [3], secretion of exotoxins [4], and multiple intrinsic and acquired resistance mechanisms [5]. Collectively, these attributes contribute to its classification as multidrug-resistant (MDR) [6], critically important [7], extensively drug-resistant (XDR), pandrug-resistant (PDR) [8], and difficult-to-treat-resistant (DTR) [9].
The emergence of resistance in P. aeruginosa occurs through de novo mutations [10,11], antibiotic-induced selection pressure, horizontal transmission between patients [12,13], and cross-resistance mechanisms, ultimately leading to MDR phenotypes [14]. In India, a scoping review reported a high prevalence of carbapenem resistance (40–47%), with more than half of the isolates additionally resistant to fluoroquinolones and third-generation cephalosporins [15]. Other studies documented MDR prevalence rates ranging from 31.78% to 50%, with some strains displaying XDR phenotypes and carbapenem resistance as high as 80% [5,16]. A recent antimicrobial resistance (AMR) survey of 58 P. aeruginosa isolates from patients in Kerala showed that 74.1% were MDR, while 55.8% were resistant to the entire panel of antibiotics tested. Alarmingly, 93% of isolates demonstrated resistance to both levofloxacin and meropenem [17].
Despite the availability of advanced antibacterial agents, the limited development of novel antibiotics underscores the urgent need for alternative therapeutic options for P. aeruginosa infections. Bacteriophages, being self-replicating bacterial viruses, represent one such promising strategy.
Phage therapy offers several advantages, including high specificity, activity against antibiotic-resistant bacteria, and minimal disturbance of the host microbiota [18]. Nevertheless, it is not without challenges: a narrow host range, potential emergence of phage-resistant mutants, and regulatory constraints remain significant barriers [19]. For therapeutic application, an ideal phage should demonstrate a strictly lytic life cycle, stability under physiological conditions, and broad activity against clinically relevant isolates [20]. Comprehensive characterization—encompassing host range determination, adsorption rate, burst size estimation, one-step growth dynamics, and genomic analysis to exclude undesirable genes such as those encoding toxins or lysogeny-associated elements—is critical to ensure phage safety, efficacy, and therapeutic suitability [21,22].
In this study, we report the isolation of novel bacteriophages from lake and sewage samples collected in Thane and Navi Mumbai, Maharashtra, India. The isolated phages were characterized for their stability, burst size, and host range, and the lytic efficacy of five selected phages was demonstrated in various in vitro models. Furthermore, we established and employed two alternative in vitro cell-based models as substitutes for animal experimentation to evaluate the antibacterial efficacy of bacteriophages. These models also hold potential for assessing the activity of other antibacterial agents.
In future work, the phages that exhibited strong antibacterial activity in vitro will undergo detailed genetic characterization to gain insights into their genome organization and functional attributes. In addition, their therapeutic potential will be further assessed in vivo using appropriate animal models to establish safety and efficacy.
2. Materials and Methods
The study was approved by the Institutional Biosafety Committee (IBSC) during its 62nd meeting held on 18 June 2018. All experimental procedures were carried out under BSL-2 conditions within a Class II Biosafety Cabinet to ensure appropriate safety measures.
2.1. Collection of Samples, Isolation and Purification
Specimens were collected from lakes and sewage systems in Thane and Navi Mumbai. Lake water was obtained from the surface, while sewage samples were collected from both the top layer and the bottom sediment. The samples were stored at 4 °C until processing [23]. For phage isolation, samples were settled for one hour, centrifuged at 2400× g for 20 min, and the supernatant was filtered through 0.22 µm syringe filters. The filtrate was used for the isolation of lytic bacteriophages against Pseudomonas aeruginosa PAQ609 by incubating 1 mL of the supernatant with 50 mL of nutrient broth containing the host bacteria at 37 °C for 24 h. After incubation, the suspension was serially diluted and plated using the double-layer agar method. Plaques were isolated, purified by repeating the process twice until homogeneity, and the phages were enriched using the host bacterium. The lysate was clarified by centrifugation at 13,000× g for 10 min, followed by filtration through a 0.22 µm Millipore membrane, and subsequently stored at −80 °C for further use.
2.2. Bacterial Strains
All P. aeruginosa strains, except MTCC 464, were provided by Wockhardt Research Centre, Aurangabad. Upon receipt, all cultures were propagated in trypticase soya broth (TSB) at 37 °C with shaking at 120 rpm. The cultures were then aliquoted into 1.5 mL cryotubes with 20% (v/v) glycerol and stored at −80 °C.
2.3. Determination of Plaque Morphology
To determine plaque morphology, serial ten-fold dilutions of bacteriophage lysate were prepared and processed using the double-layer agar method. After 24 h of incubation at 37 °C, individual plaques were analyzed for morphology, including clear centers with turbid edges and halos, clear plaques, and bull’s eye-shaped plaques. Plaque size was measured using a Vernier caliper.
2.4. Electron Microscopy
A phage lysate with a concentration of 1010 PFU/mL was passed through a 0.22 µm syringe filter and stored at 2–8 °C. Subsequently, 1 mL of the filtered lysate was centrifuged at 11,000× g for 2 h at 25 °C. The supernatant was carefully removed, and the resulting pellet was incubated in 5 mM MgSO4 solution at 2–8 °C for 24 h to facilitate resuspension. Following incubation, the pellet was gently resuspended by pipetting.
For electron microscopy analysis, the phage suspension was mixed with 4% paraformaldehyde in a 1:1 ratio for fixation. Fixed phage particles were applied onto coated copper grids and allowed to adsorb for 10 min. Grids were then negatively stained with 1% aqueous uranyl acetate for 10 s and air-dried for 2 h. Imaging was performed using a JEM 1400 Plus transmission electron microscope (JEOL, Tokyo, Japan) operated at 120 kV.
2.5. One Step Growth Curve and Determination of Burst Size
The one-step growth experiment was carried out as described in [24] with minor modifications. Briefly, a bacteriophage suspension (final concentration: 1 × 107 PFU/mL) was mixed with a bacterial culture adjusted to 1 × 109 CFU/mL, corresponding to a multiplicity of infection (MOI) of 0.01. The mixture was incubated at 37 °C for 8 min to allow adsorption, followed by centrifugation at 13,000× g for 5 min. The supernatant was discarded, and the pellet was resuspended in 100 mL of nutrient broth. Aliquots were collected at 5-min intervals over a period of 100 min, serially diluted, and plated with the host strain using the double-layer agar method for phage enumeration. Each experiment was performed in triplicate. The latent period was defined as the interval between the completion of adsorption and the initial rise in phage titers, indicating the onset of lysis. Burst size was calculated as the ratio of the total number of phage progeny released to the number of infected bacterial cells during the latent period.
2.6. Temperature and pH Stability
The thermal stability of bacteriophages (5 × 106) was assessed at temperatures of 4 °C, 25 °C, 37 °C, 45 °C, 50 °C, and 60 °C in nutrient broth at pH 7.4 for one hour in a pre-adjusted water bath. For pH stability, the phages were tested at pH levels of 2, 3, 4, 5, 6, 7, 8, 10, 12, and 13 at 37 °C, with pH adjustments made using 6 M HCl or 6 M NaOH. After incubation, surviving bacteriophages were counted using the double-layer agar method.
2.7. Minimum Inhibitory Concentration of Antibiotics
To determine antibiotic resistance, all P. aeruginosa strains were subjected to broth microdilution MIC following CLSI guidelines [25]. A panel of 14 antibiotics from various classes, including fluoroquinolones, macrolides, and carbapenems, was used. Antibiotic stocks were prepared in the appropriate solvents, diluted in Muller Hinton broth, and added to a 96-well plate along with 5 × 105 bacterial cells per well. Positive and negative controls were included. Plates were incubated at 37 °C for 24 h, and MIC was recorded as the lowest antibiotic concentration with no visible bacterial growth.
The MIC of bacteriophages was determined using the broth microdilution method by Vipra et al., 2013 [26]. Phages were serially diluted in Muller Hinton broth (MOI 0.001-1000) and added to 96-well plates with 5 × 105 P. aeruginosa PAQ609 cells per well. Each phage MOI was tested in triplicate. Positive controls contained sterile media, and negative controls had media with bacteria. Plates were incubated at 37 °C for 24 h, and the minimum inhibitory phage concentration was identified as the lowest MOI with no visible bacterial growth.
2.8. Phage Host Range Determination
The host range of phages against P. aeruginosa was determined using a spot assay as described by Khan et al., 2015 [27]. Soft agar mixed with 100 µL of log-phase bacterial culture was poured onto nutrient agar plates. After solidification, 10 µL of 1 × 109 phage suspensions were spotted onto the plates. Following 24-h incubation at 37 °C, bacterial sensitivity was assessed by observing the lysis zone. Strains showing lysis were marked as susceptible (‘+’), while those without lysis were marked as resistant (‘−’).
2.9. Time Kill Assay
The time-dependent bactericidal activity of bacteriophages against planktonic Pseudomonas aeruginosa was investigated. An initial bacterial suspension of 108 CFU/mL was prepared, and phage treatments were applied at multiplicities of infection (MOIs) of 0.1, 1, and 10, both individually and as a combined phage cocktail (PA1–PA5). Phage-free cultures served as negative controls, while ciprofloxacin at 2× MIC was used as a positive control. All samples were incubated at 37 °C with continuous agitation for up to 48 h. Bacterial growth inhibition was monitored by measuring optical density at 600 nm at specific time points (0, 2, 4, 6, 24, and 48 h) [28].
2.10. Inhibition of Biofilm
The anti-biofilm activity of bacteriophages against established P. aeruginosa PAQ609 biofilms was evaluated following the methods of O’Toole (2011) [29] and Lalitha et al. (2017) [30]. P. aeruginosa biofilms were grown in 96-well plates for 48 h at 37 °C. After removing unattached cells, bacteriophage suspensions (108 and 109 PFU/mL) were added for 6, 24, and 48 h. Both single-phage applications and a phage mixture (PA1–PA5) were tested. Biofilms treated with ciprofloxacin (2× MIC) served as positive controls, while untreated biofilms were negative controls. Biofilm inhibition was assessed via crystal violet staining, and optical density was measured at 590 nm. The percentage reduction of biomass was determined using the following formula:
Percent total biomass = OD of cultures with phage/OD of cultures without phage (Negative control) × 100
Percent biomass inhibition = 100 − % total biomass
Biofilm viability was assessed using the MTT assay, which detects metabolically active cells. After incubation, the supernatant was discarded, and 0.1 mL nutrient broth and 0.01 mL MTT (5 mg/mL) were added to each well. Plates were incubated for 2 h at 37 °C, followed by the addition of 0.1 mL dimethyl sulphoxide to dissolve formazan crystals. Absorbance was measured at 570 nm, with 630 nm as the reference, using an ELISA plate reader (PowerWave XS microplate spectrophotometer, BioTek, VT, USA).
Percent viability was determined using the following formula:
OD of cultures with phage or antibiotic/OD of cultures without phage or antibiotic (Negative control) × 100
2.11. Anti-Biofilm Efficacy
Bacteriophage penetration through biofilm was assessed using the method by Singh et al., 2010 [31]. A biofilm of P. aeruginosa was formed on 10 mm nitrocellulose disks (0.4 µm pore size) and incubated on nutrient agar plates at 37 °C for 48 h. These disks were then placed on pre-seeded nutrient agar plates with the host bacteria, and 5 mm nitrocellulose disks loaded with 0.01 mL of 109 PFU/mL phage suspension were applied on top. After a 24-h incubation at 37 °C, the zone of inhibition around the 10 mm disk was measured, indicating phage penetration through the biofilm.
2.12. In Vitro Wound Infection Model—Scratch Assay
The mechanical wound healing assay was performed using a modified protocol based on Liang et al., 2007 [32]. L929 fibroblast cells (ATCC) were maintained in Minimum Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 atmosphere. After reaching confluence, a uniform scratch was generated in the cell monolayer using a sterile micropipette tip, and detached cells were removed by washing with phosphate-buffered saline (PBS). The wells were then replenished with MEM containing 10% FBS and infected with Pseudomonas aeruginosa at a concentration of 1 × 106 CFU/mL for 3 h. Following bacterial exposure, the medium was discarded, and a bacteriophage cocktail (1 × 109 PFU/well) was applied. Appropriate positive and negative controls were included. Plates were incubated for 24 h, after which cells were fixed using 70% acetone and stained with 1% crystal violet. Wound closure was documented at 0, 24, 48, and 72 h using an inverted microscope (Zeiss India, Bengaluru, India), and image analysis was performed with ImageJ software (National Institute of Health, Bethesda, Maryland, USA; Ver 1.54e) to quantify the percentage of wound closure.
2.13. In Vitro Burn Wound Infection Model
The in vitro burn infection model was established based on Fernandes et al., 2014 [33], with adaptations. L929 fibroblast cells were cultured in LabTek chamber slides and allowed to adhere overnight. Prior to burn induction, cells were trypsinized and counted using the trypan blue exclusion assay. Thermal injury was applied by exposing the chambers to 70 °C for 18 s, after which the medium was immediately replaced with MEM containing 10% FBS at 4 °C to arrest further thermal damage. Following the burn, cells were infected with Pseudomonas aeruginosa at a concentration of 1 × 106 CFU/mL and incubated at 37 °C with 5% CO2 for 3 h. Subsequently, the medium was replaced with either a bacteriophage cocktail (1 × 109 PFU/well) or ciprofloxacin (4 µg/mL), and all conditions were prepared in triplicate. Negative controls consisted of infected but untreated cells, while positive controls were uninfected. The cultures were maintained for 72 h, with one chamber collected every 24 h to determine viable cell counts using trypan blue exclusion.
3. Results
3.1. Isolation of Bacteriophages, Plaque Morphology and Electron Microscopy
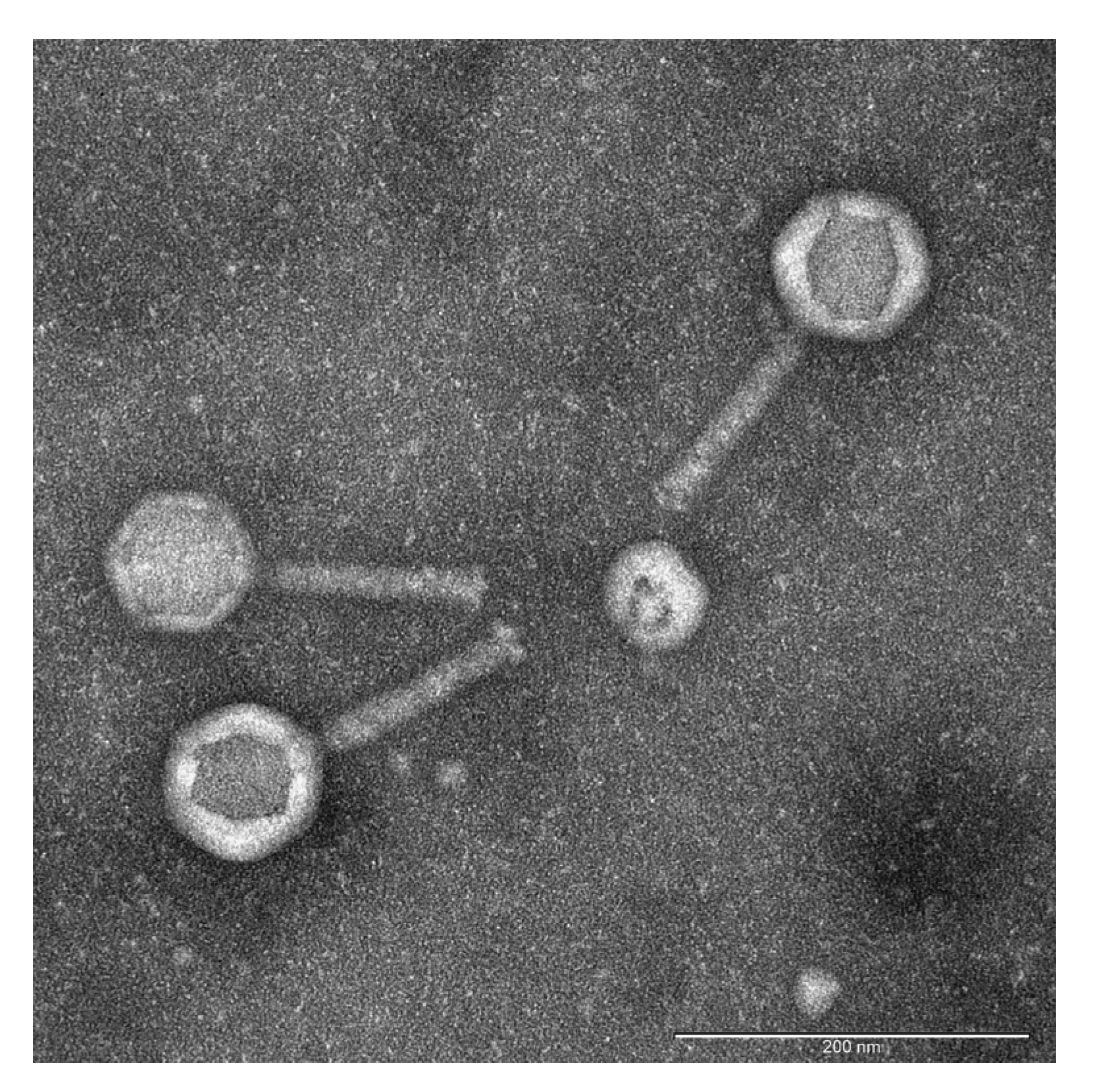
To isolate novel bacteriophages active against Pseudomonas aeruginosa, environmental samples of water and sewage were collected from lakes and wastewater outlets in Thane and Rabale, Navi Mumbai, Maharashtra, India (Supplementary Table S1). Following enrichment and serial dilution, distinct plaques were identified, with some exhibiting clear and others translucent morphologies. Plaques with clear zones were selected, resulting in the isolation of 13 phages. The plaque sizes ranged from 1.5 mm to 2.5 mm, all displaying clear centers with turbid halos. These phages were designated as PA1 through PA13. Transmission electron microscopy (TEM) of the most potent phage, PA2, revealed morphological characteristics consistent with Myoviruses: a contractile tail measuring 118 ± 4 nm and an icosahedral head of 88 ± 8 nm. Consequently, PA2 was renamed Pseudomonas phage vB_PaeM-PA2 [34] (Figure 1).
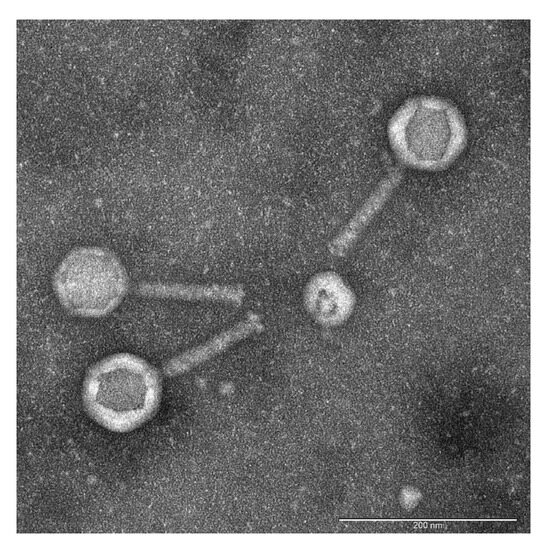
Figure 1.
Transmission electron micrograph of negatively stained bacteriophage PA2. The scale bar represents 200 nm. Phage PA2 exhibited a contractile long tail measuring 118 ± 4 nm in length, with an overall length of 212 nm. Morphological analysis by TEM revealed structural features characteristic of Myovirus, including an icosahedral head connected to the tail tube by a distinct neck structure.
3.2. Host Range Determination
Host range testing revealed that phages PA2, PA7, PA8, PA10, and PA11 had broad host ranges, lysing 53.33% (8/15) of the tested multi-drug resistant Pseudomonas aeruginosa strains. Phages PA1, PA4, PA9, and PA12 lysed 46.66% (7/15) of the strains, while PA3, PA5, and PA6 were effective against 40% (6/15). Phage PA13 exhibited a restricted host range, lysing only 3 out of 15 strains.
Based on the experiment’s results, a phage cocktail consisting of PA1, PA2, PA3, PA4, and PA5, each with different host ranges, was created to assess if it could expand the number of sensitive hosts. Though PA2 and PA5 exhibited identical host ranges, both were still included in the cocktail formulation. This combination successfully lysed 9 out of 15 (60%) multi-drug-resistant Pseudomonas aeruginosa strains. Consequently, all subsequent experiments focused on these five phages (Table 1, Supplementary Figure S2).

Table 1.
Host range determination for Pseudomonas-specific phages.
3.3. One Step Growth Curve
This experiment measured the latent time and burst size per infected bacterial cell for Pseudomonas aeruginosa-specific phages. The latent period ranged from 35 to 50 min, averaging 42 ± 8 min. Phages PA2 and PA3 had the longest latent periods at 50 min, while PA4 and PA5 had the shortest at 35 min. Burst sizes varied, with PA2, PA4, and PA5 exhibiting the largest burst sizes (103, 163, and 198 PFU/ cell, respectively), whereas PA1 and PA3 had the smallest (76 and 22 PFU/cell). The average burst size was 112 ± 70 PFU/cell.
3.4. pH and Thermal Stability
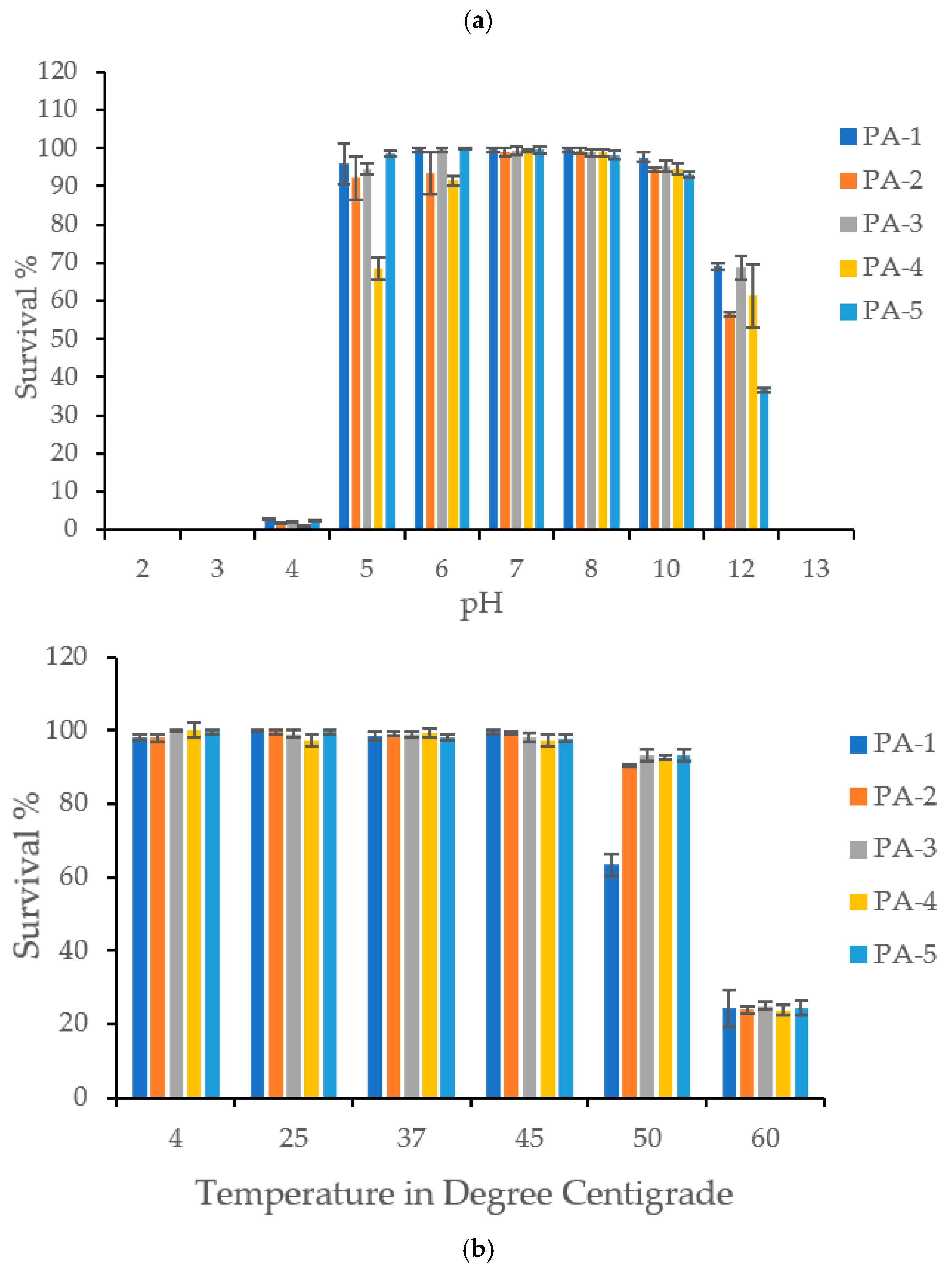
Phages maintained comparable viability and stability at pH levels ranging from 5 to 11. However, at pH 12, viability significantly decreased by 31–64%, and at pH 13, phages completely lost their stability and viability. The phages were also unstable in acidic conditions (pH 2, 3, and 4) but retained 68–98% viability at pH 5 (Figure 2).
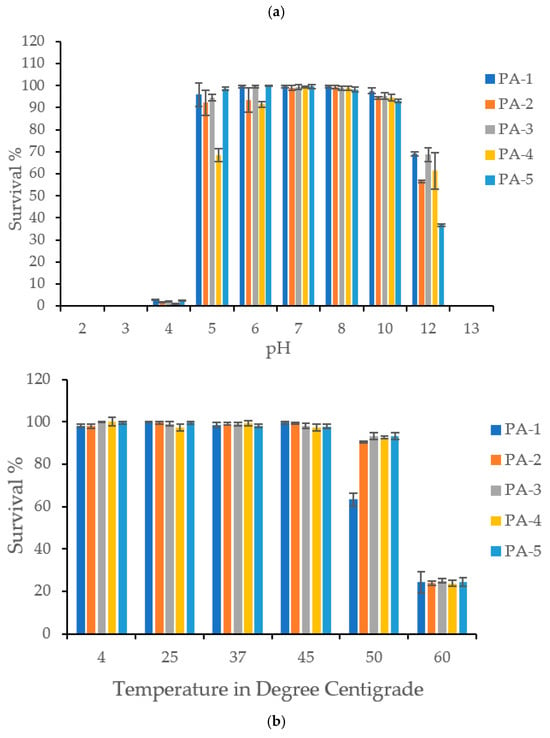
Figure 2.
(a) pH Stability: The figure shows stability of P. aeruginosa specific phages PA1, PA2, PA3, PA4, and PA5 at various pH ranging from 2–13. Phages were found stable from pH 5–12. All the phages demonstrated complete loss of viability at pH 13. All the phages were found very sensitive to acidic pH such as 2, 3 and 4. (b)Thermal Stability: The figure shows thermal stability of P. aeruginosa specific phages PA1, PA2, PA3, PA4, and PA5 at various temperatures. Phages were found stable at temperature range of 4 °C to 50 °C. PA2, PA3, PA4, and PA5 demonstrated significant loss in viability at 60 °C. PA1 exhibited its thermal stability from 4 to 45 °C, however a sharp drop was observed at 45 °C.
In terms of temperature stability, the phages remained stable across a broad range from 4 °C to 50 °C. However, at 60 °C, they became unstable, losing an average of 75% viability (Figure 2).
3.5. Minimum Inhibitory Concentration of Antibiotics
All Pseudomonas aeruginosa strains tested were resistant to ampicillin, amoxicillin, augmentin, and cefixime. About 90% of the strains showed resistance to azithromycin, aztreonam, and ceftriaxone, while 80% were resistant to cefixime. Resistance to fluoroquinolones, such as ciprofloxacin and levofloxacin, was observed in approximately 66% of the strains. Meropenem resistance was found in about 26% of the strains (Supplementary Table S2).
3.6. Minimum Inhibitory Concentration for Phages
When phages were incubated at varying multiplicities of infection (MOI) from 0.001 to 1000 with a fixed number of P. aeruginosa PAQ609 cells (5 × 105/well), they demonstrated significant inhibitory activity within 24 h. The minimum inhibitory concentration (MIC) of the phages ranged from 0.07 ± 0.052 to 1.0 ± 0.0 MOI. Phages PA2 and PA3 had the lowest MIC (0.07 ± 0.052 MOI), while PA1 and PA5 had the highest (1.0 ± 0.0 MOI). Phage PA4 had a MIC of 0.7 ± 0.52 MOI (Supplementary Table S3).
3.7. Time Kill Assay
Bacterial growth dynamics were monitored by measuring optical density at regular intervals during the time-kill assay under continuous agitation. At all evaluated MOIs, individual and combined treatments with phages PA1, PA2, PA3, PA4, and PA5 resulted in a marked reduction in optical density after 8 h of incubation.
All phages at all MOIs achieved an 80% reduction in optical density after 8 h. Phages PA1, PA2, and PA3 were particularly bactericidal, showing a significant decrease in optical density. However, phages PA1 (MOI 10), PA2 (MOI 1 and 10), and PA3 (MOI 1 and 10) exhibited regrowth by the 24-h mark, indicated by no further reduction and an increase in optical density.
Although PA4 and PA5 were initially bactericidal, regrowth was observed after 24 h. Ciprofloxacin (4 µg/mL) reduced cell density by 78% after 8 h, but bacterial regrowth occurred by 24 h. However, at 48 h, a combination of phages PA1, PA2, PA3, PA4, and PA5 effectively lysed the bacterial host and prevented regrowth (Supplementary Figure S1).
3.8. Biofilm Inhibition and Determination of Antibiofilm Efficacy
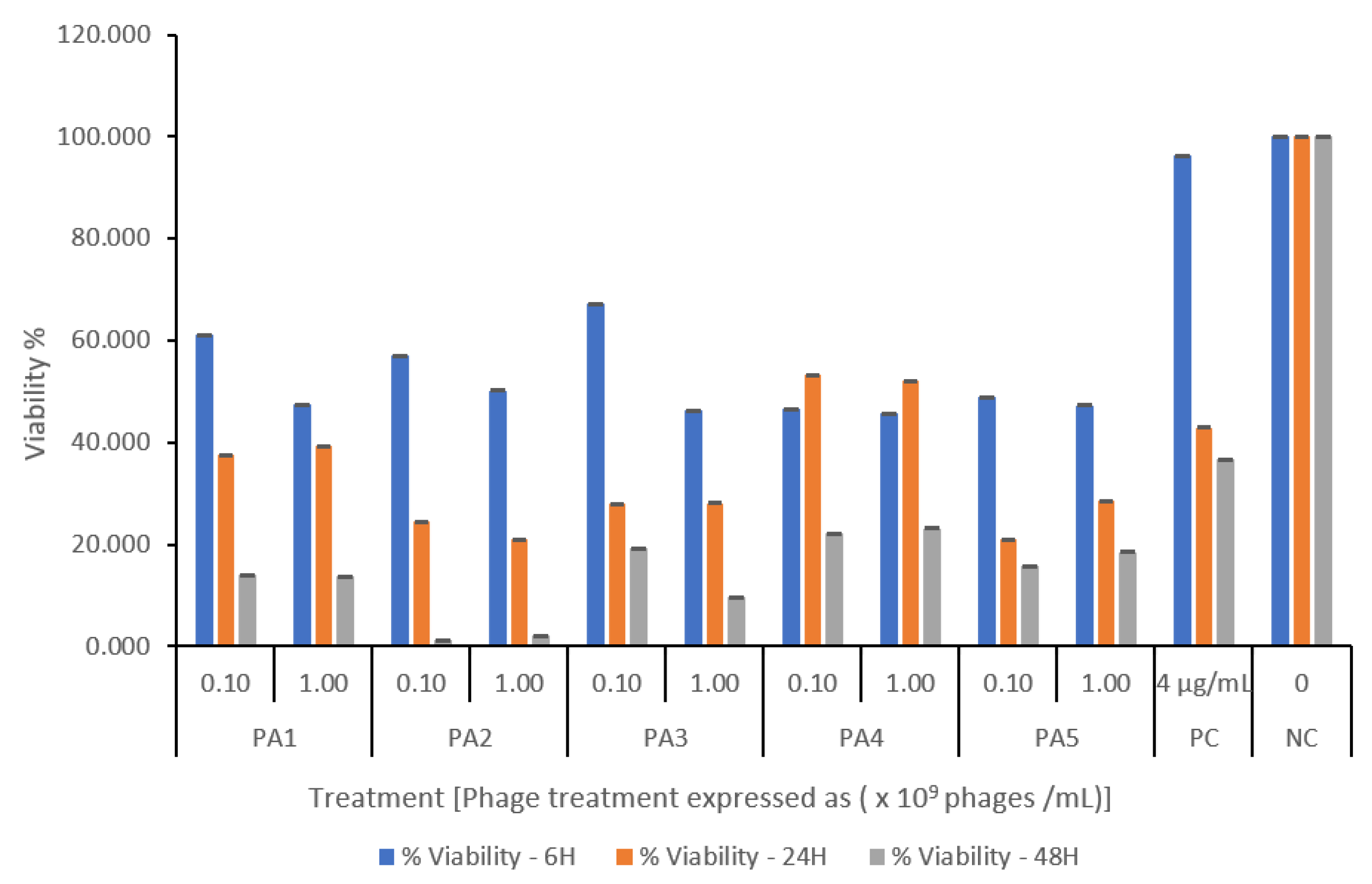
All bacteriophage-treated cultures showed a titer- and time-dependent reduction in biomass when applied to established biofilms. Phage PA1 (109) was the most effective, reducing biomass by 80% after 48 h. Except for PA3, all phages reduced biomass viability to below 30% at all titers. Phages acted rapidly on the biofilm, causing about a 30% reduction in viability within just 6 h, whereas ciprofloxacin (4 µg/mL) was only effective after 48 h (Figure 3).
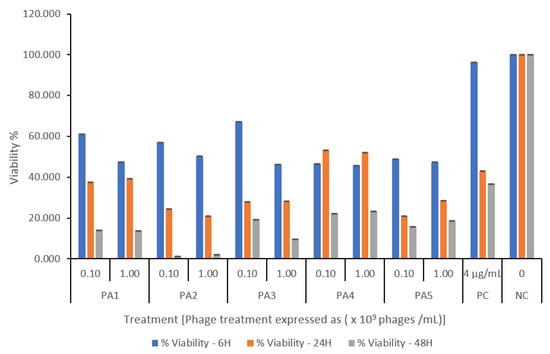
Figure 3.
Pseudomonas aeruginosa-specific phage-mediated biofilm inhibition assessed by MTT assay. Biofilms were treated with phage at two inoculum levels (108 and 109 PFU/mL). Untreated biofilms served as negative control, and ciprofloxacin (4 µg/mL) was used as positive control. Data are presented as percent viability relative to control, expressed as mean ± SD of triplicate wells. A significant reduction in biofilm viability was observed in all phage-treated cultures within 6 h of treatment compared to the untreated control. Phage treatment continued to suppress biofilm, achieving ≤ 30% viability by 48 h.
Additionally, all phages were evaluated for their anti-biofilm efficacy by assessing their ability to penetrate biofilms, with lytic zones ranging from 10.57 ± 0.23 to 13.57 ± 0.06 mm. Phage PA5, however, exhibited significantly lower anti-biofilm efficacy, as indicated by the smallest zone of inhibition compared to the other phages (Supplementary Table S4).
3.9. In Vitro Burn Wound Infection Model
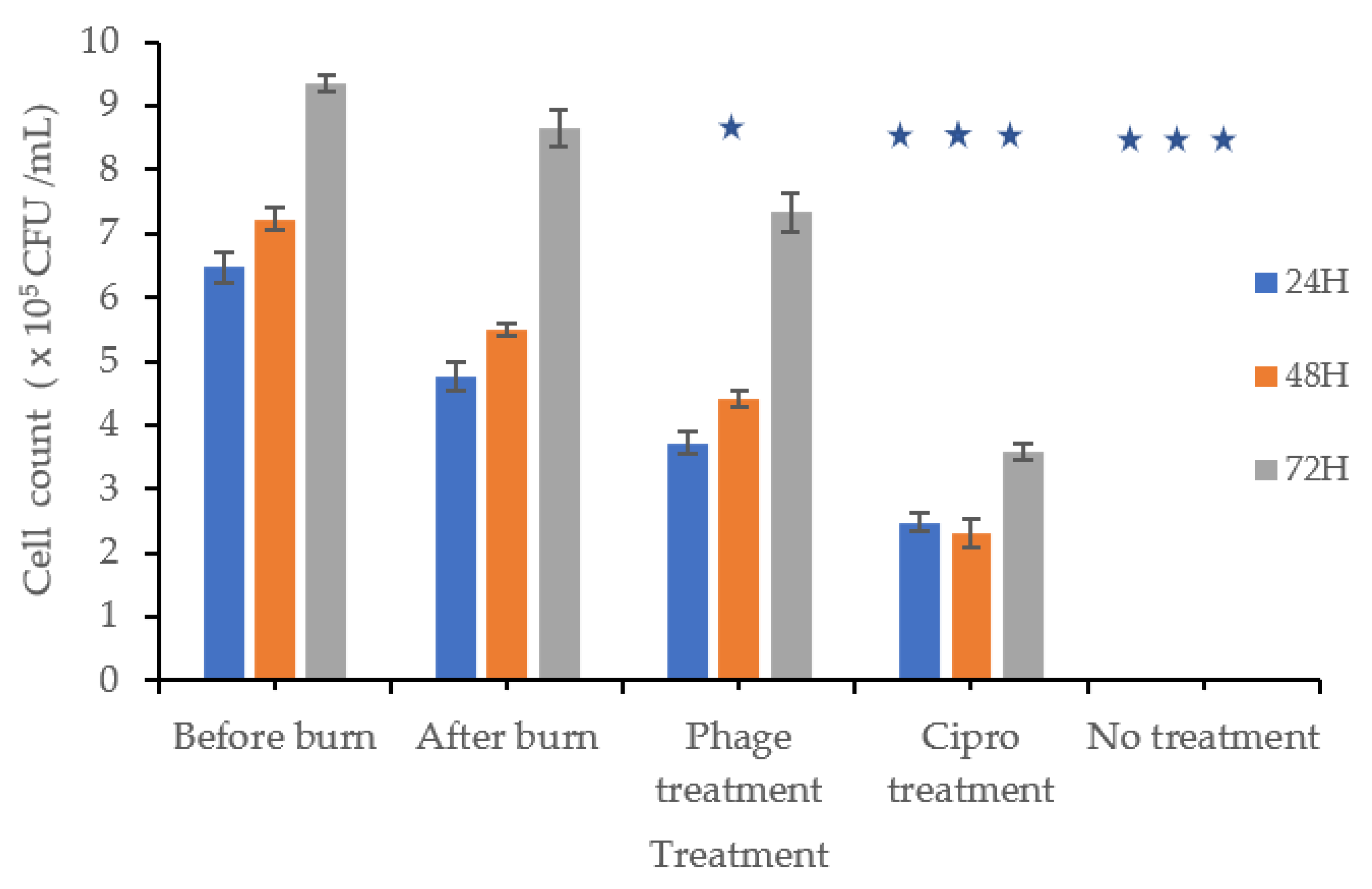
Cells were counted before and after burn injury, exposed to dry heat, and then treated with a phage mixture, a positive or negative control, or ciprofloxacin. In the positive control group, cell counts increased by 1.29, 1.49, and 2.34 times after 24, 48, and 72 h, respectively. In contrast, cells treated with the phage mixture multiplied by 1.01, 1.19, and 1.98 times over the same time periods (Figure 4). Although cell proliferation was observed in the phage-treated wells, it was less pronounced than in the positive control.
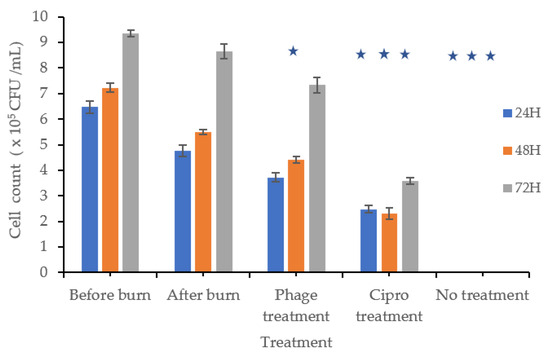
Figure 4.
Burn Wound Infection: The bars labeled “Before Burn” represent L929 cell counts at 24, 48, and 72 h in unburned, uninfected cultures to monitor normal cell growth over time. The “After Burn” bars represent cell counts at the same time points after inducing a burn, but without bacterial infection, to evaluate the effect of burn injury on cell growth. Subsequently, burned L929 cells were infected with P. aeruginosa and treated with either a phage mixture, ciprofloxacin, or left untreated. Cell viability was measured at 24, 48, and 72 h post-treatment to assess cell proliferation. At 48 h, the phage-treated group showed significantly lower cell proliferation compared to the uninfected control (p ≤ 0.001), while cell counts partially recovered by 72 h, although they remained lower than the uninfected control. These findings indicate that phage treatment effectively controlled bacterial infection and supported partial recovery of L929 cells. Cultures treated with ciprofloxacin or left untreated showed significantly lower cell counts at all time points (p ≤ 0.001). Statistical analysis was performed using T-test. Statistical significance (p ≤ 0.001) is indicated in the graph with star (*).
Wells infected with P. aeruginosa and treated with ciprofloxacin (4 µg/mL) showed a fold change of 0.67, 0.62, and 0.96 after 24, 48, and 72 h, respectively.
3.10. In Vitro Wound Infection Model—Scratch Assay
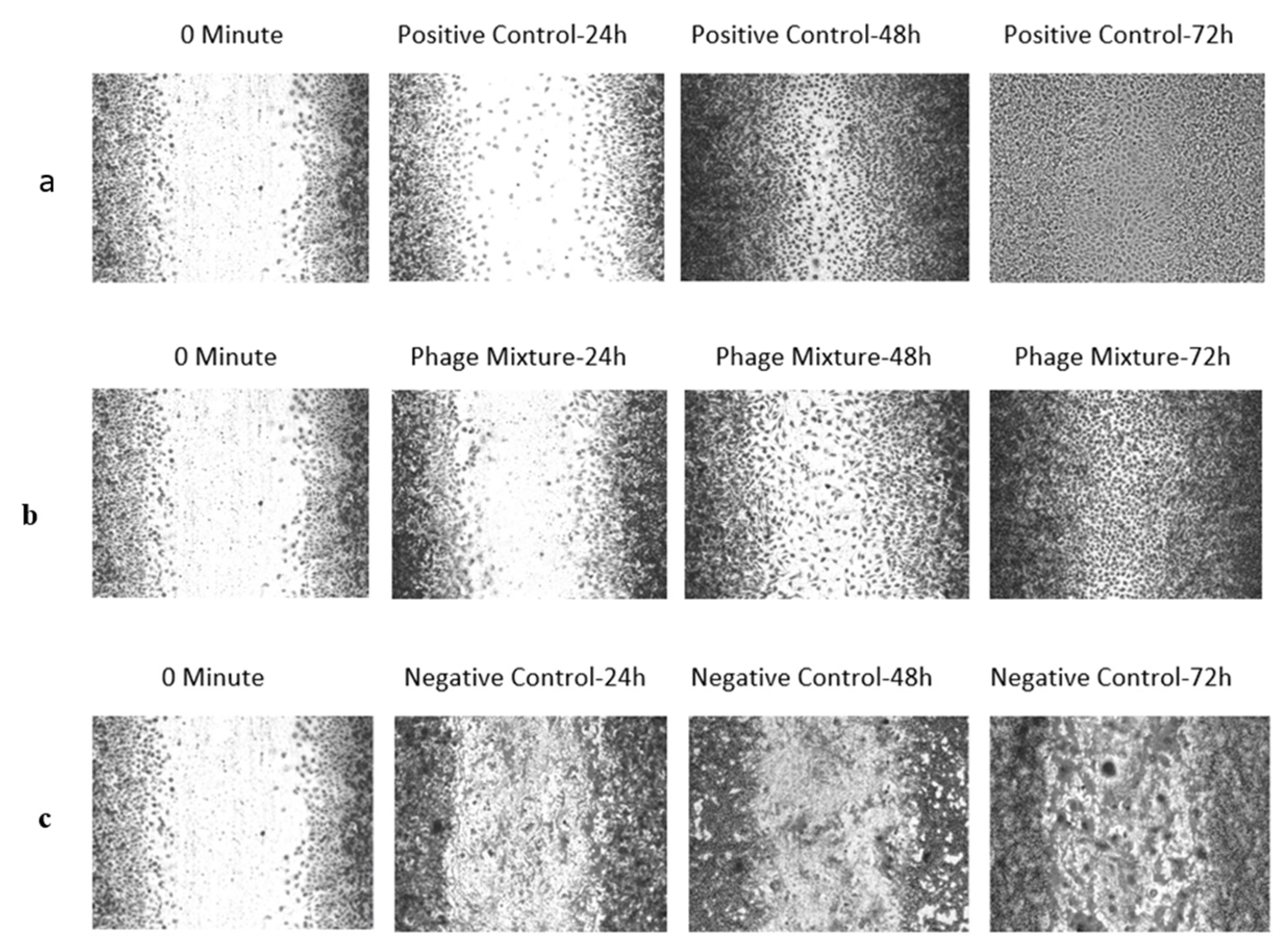
In this 2D model of mechanical wound infection, infected wounds were treated with a phage mixture, and wound closure was monitored at 0, 24, 48, and 72 h. The wound gap area was measured using ImageJ software, with the mean of ten measurements used as the average remaining gap.
The phage cocktail consistently inhibited bacterial proliferation throughout the experiment, while enhancing cell viability, proliferation, and migration within the wound region. The positive control achieved complete wound closure after 48 h. After 24 and 48 h, phage treatment resulted in a statistically significant (p < 0.05) reduction in wound gap compared to positive control cultures. Cell proliferation and migration were observed in the phage-treated wounds by 72 h, leading to gap closure, although not to the extent seen in the positive control (Figure 5).
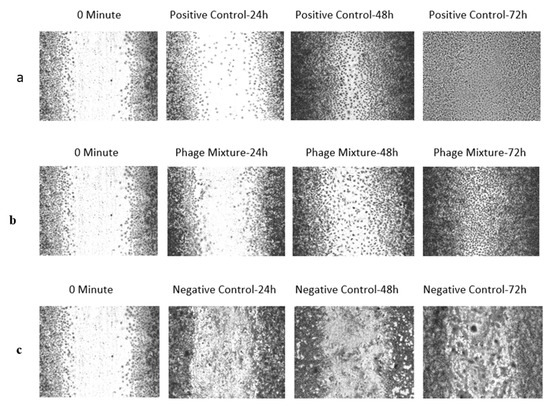
Figure 5.
In vitro wound infection model—scratch assay illustrating the effect of Pseudomonas aeruginosa infection and phage treatment on L929 cell proliferation and migration. (a) Un-Infected control—Wounds without P. aeruginosa infection showed normal L929 proliferation and migration, resulting in complete closure of the wound gap after 72 h. (b) Treatment with phages—Wounds infected with P. aeruginosa and treated with a phage mixture exhibited inhibition of bacterial growth, allowing partial L929 proliferation and migration; the wound gap was completely closed by 72 h, although closure was not as extensive as in the positive control. (c) Negative control (Untreated Control), in which wounds were created but left untreated, showed biofilm formation within the wound gap, with cells losing their normal morphology and appearing rounded.
4. Discussion
Antibiotic resistance is a growing global concern, potentially causing 10 million deaths annually by 2050 [35]. The improper and widespread use of antibiotics has led to multi-drug resistance (MDR) in bacteria, particularly Gram-negative strains [36], posing a major challenge in developing effective treatments. To combat this, alternatives like phage therapy, or combining antibiotics with adjuvant therapies, are being explored. Phage therapy, despite some limitations, is gaining traction due to its specificity for bacterial hosts, self-replication, simple isolation, and low production costs, making it a promising alternative to antibiotics.
In a recent study, Pseudomonas aeruginosa PAQ609, which is resistant to penicillins, macrolides, tetracyclines, monobactams, and third-generation cephalosporins, was used as the host for bacteriophage isolation. Thirteen phages were successfully isolated from 16 environmental samples.
The isolated phages exhibited diverse plaque morphologies, all featuring clear centers surrounded by turbid edges and halos. The halos are thought to result from phage-produced depolymerase enzymes [37], which degrade biofilms and facilitate antibiotic delivery, making them therapeutically valuable. The halos are also a hallmark of tail spike protein (TPS) activity, characteristic of capsule-targeting phages [38]. After establishing their morphological characteristics, we tested the phages for burst size and tolerance to physiological variables like pH and temperature. Unlike antibiotics, phage therapy benefits from being a self-replicating treatment, ideally with a short latent period and large burst size to quickly inactivate target bacteria. Our investigation revealed diverse growth patterns among the phages, with variations in burst size and latent periods, indicating a wide genetic diversity in the environment. This experiment also differentiated phages with similar plaque morphologies based on their latent time and burst size.
Literature indicates that rising temperatures increase bacterial antibiotic resistance [39], while pH changes affect antibiotic efficacy [40]. An effective therapeutic agent must retain its potency despite temperature and pH fluctuations in vivo. Bacteriophages, when produced in large quantities, may need to withstand temperature and pH changes during synthesis, storage, transport, and administration. For optimal therapeutic effectiveness, phages must endure wide pH and temperature variations and be present in sufficient numbers to rapidly eliminate the target. Infected wounds [38] and urine in urinary tract infections are reported to be alkaline [39], as is the pH of healthy lung airways [40]. Phages used therapeutically against P. aeruginosa must remain stable at alkaline pH, as these bacteria are associated with infections in wounds, lungs, and the urinary system. Given that the human body temperature is 37 °C, phages must also maintain stability at this temperature and slightly higher. Our study found that all tested phages exhibited optimal viability and stability across a wide pH range (5–12) and temperature range (4–50 °C). In contrast, antibiotics like penicillin G, ampicillin, cephalothin, cephaloridine and novobiocin are more effective at pH 5.0 than in more alkaline media [41], limiting their use in lung, wound, and urinary tract infections. In such cases, phages could be effectively used as adjuvant therapy.
To effectively treat bacterial infections, we need better therapeutic agents and strategies, requiring careful selection of antimicrobials based on microbiological parameters. Host range determination is a key factor in selecting phages for therapy. In our study, many phages were effective against multi-drug resistant, ESBL-producing, and carbapenemase-producing strains, making them strong candidates for phage therapy. Based on these findings, a cocktail of five broad-spectrum phages was developed. One phage with a broad host range was selected for electron microscopy. Morphological features are crucial for phage identification. Transmission electron microscopy revealed that phage PA2 possesses an icosahedral head connected to a tail tube by a neck, a morphology characteristic of Myoviruses.
In the time-kill assay, phages rapidly targeted their hosts, exhibiting lytic activity regardless of MOI. All phages tested against Pseudomonas showed a significant reduction in cell density within just two hours of infection, supporting the concept of self-replicating medicine. Phage combinations prevented bacterial regrowth, indicating that these phages target different host surface receptors. As a result, even if the bacteria developed resistance to one phage, the others would still control the infection. This highlights the therapeutic value of using a phage mixture, as it reduces the likelihood of resistance development.
Cells embedded within biofilms display greater mechanical stability than their planktonic counterparts, which contributes to their enhanced resistance against antimicrobial agents [42]. Application of the phages led to a substantial reduction in biofilm biomass accompanied by a significant decline in bacterial viability. The halos around the clear centers of nearly all phages against P. aeruginosa indicate the production of depolymerases, which contribute to their rapid degradation ability.
Several classes of antibiotics have been found unable to penetrate S. aureus and S. epidermidis biofilms, contributing to drug resistance [31]. Given this, we investigated whether phages could overcome this limitation. Our findings revealed that nearly all Pseudomonas-specific phages could cross the exopolysaccharide barrier of biofilms and exert antimicrobial effects, highlighting their significant therapeutic potential.
Open wounds are highly susceptible to opportunistic pathogens due to poor blood flow and a hypoxic environment that fosters bacterial growth. Bacteria form multicellular biofilms on the wound bed, making infections difficult to treat [43]. In this study, we explored the use of bacteriophages to treat wound infections in a 2D model. The phage combination successfully inhibited bacterial growth while supporting host cell proliferation and migration within the wound site. After 72 h, the wound gap was fully closed despite initial delays caused by the bacterial infection, which had impeded cell growth before the phages took effect.
In this study, we also developed a burn wound infection model. Burns require urgent medical attention as the burn environment attracts and fosters the growth of both Gram-positive and Gram-negative bacteria. Bacterial contamination in severe burn wounds presents a significant medical challenge, with P. aeruginosa being the most dangerous Gram-negative pathogen. It can severely delay healing and lead to fatal outcomes [44].
In this study, time-lapsed cell counts showed that P. aeruginosa significantly reduced cell numbers at all time points compared to uninfected controls, confirming its detrimental impact on burn wounds and delayed healing. Phage-treated cultures were more effective than ciprofloxacin treatment. Based on these in vitro models and experimental results, we conclude that these phages are superior candidates for therapy. This study also demonstrated that modified in vitro animal alternative models can be effectively used to assess the efficacy of antibacterial agents.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomed5040025/s1, Figure S1: In vitro time-kill assay evaluating the lytic activity of a phage cocktail (PA1, PA2, PA3, PA4, and PA5) against Pseudomonas aeruginosa; Figure S2: Host specificity of Pseudomonas aeruginosa-specific bacteriophages against multidrug-resistant (MDR) isolates; Table S1: Source of the sample and phages isolated from the samples against P. aeruginosa; Table S2: Minimum inhibitory concentrations of selected antibiotics for various strains of P. aeruginosa; Table S3: Minimum inhibitory phage concentration for P. aeruginosa PAQ609; Table S4: The zones of lysis (mean ± SD) on nutrient agar plates seeded with P. aeruginosa PAQ609.
Author Contributions
N.S., C.S., E.A. and A.K. designed the study. N.S. conceived the study, optimized experiments, analysed data and wrote the manuscript. C.S., E.A. and A.K. critically reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was approved by the Institutional Biosafety Committee (IBSC, Reliance Life Sciences Pvt. Ltd., Mumbai, India) during its 62nd meeting held on 18 June 2018. All experimental procedures were performed within a Class II Biosafety Cabinet under BSL-2 conditions.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data generated or analysed during this study are included in this article and Supplementary Materials.
Acknowledgments
The authors acknowledge the financial support of the Deakin India Research Initiative (DIRI) program initiated between the Reliance Institute of Life Sciences, India, and Deakin University, Australia. The authors also gratefully acknowledge Reliance Life Sciences Pvt. Ltd., Mumbai, India for providing financial support for the project.
Conflicts of Interest
Nikhil Sathe is an employee of Reliance Life Sciences Pvt. Ltd. The other authors declare no conflicts of interest.
References
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence factors of Pseudomonas aeruginosa and antivirulence strategies to combat its drug resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Li, X.; Gu, N.; Huang, T.Y.; Zhong, F.; Peng, G. Pseudomonas aeruginosa: A typical biofilm forming pathogen and an emerging but underestimated pathogen in food processing. Front. Microbiol. 2023, 13, 1114199. [Google Scholar] [CrossRef] [PubMed]
- Javanmardi, F.; Emami, A.; Pirbonyeh, N.; Keshavarzi, A.; Rajaee, M. A systematic review and meta-analysis on Exo-toxins prevalence in hospital acquired Pseudomonas aeruginosa isolates. Infect. Genet. Evol. 2019, 75, 104037. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Mishra, P.P.; Premi, H.K.; Walia, A.; Dhawan, S.; Kumar, A. Increasing incidence of multidrug resistant Pseudomonas aeruginosa in inpatients of a tertiary care hospital. Int. J. Res. Med. Sci. 2014, 2, 1302–1306. [Google Scholar] [CrossRef]
- Yu, Z.; Tang, J.; Khare, T.; Kumar, V. The alarming antimicrobial resistance in ESKAPEE pathogens: Can essential oils come to the rescue? Fitoterapia 2020, 140, 104433. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Cosentino, F.; Viale, P.; Giannella, M. MDR/XDR/PDR or DTR? Which definition best fits the resistance profile of Pseudomonas aeruginosa? Curr. Opin. Infect. Dis. 2023, 36, 564–571. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; A Bonomo, R.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America 2022 guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2022, 75, 187–212. [Google Scholar] [CrossRef]
- Aloush, V.; Navon-Venezia, S.; Seigman-Igra, Y.; Cabili, S.; Carmeli, Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob. Agents Chemother. 2006, 50, 43–48. [Google Scholar] [CrossRef]
- Feng, Y.; Hodiamont, C.J.; van Hest, R.M.; Brul, S.; Schultsz, C.; ter Kuile, B.H. Development of antibiotic resistance during simulated treatment of Pseudomonas aeruginosa in chemostats. PLoS ONE 2016, 11, e0149310. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morgan, D.J.; Rogawski, E.B.; Thom, K.A.; Johnson, J.K.; Perencevich, E.N.; Shardell, M.; Leekha, S.; Harris, A.D. Transfer of multidrug-resistant bacteria to healthcare workers’ gloves and gowns after patient contact increases with environmental contamination. Crit. Care Med. 2012, 40, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Raman, G.; Avendano, E.E.; Chan, J.; Merchant, S.; Puzniak, L. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2018, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Peghin, M. How to manage KPC infections. Ther. Adv. Infect. Dis. 2020, 7, 2049936120912049. [Google Scholar] [CrossRef]
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The mortality burden of multidrug-resistant pathogens in India: A retrospective, observational study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef]
- Gill, J.S.; Arora, S.; Khanna, S.P.; Kumar, K.H. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J. Glob. Infect. Dis. 2016, 8, 155–159. [Google Scholar] [CrossRef]
- Menon, N.D.; Somanath, P.; Jossart, J.; Vijayakumar, G.; Shetty, K.; Baswe, M.; Chatterjee, M.; Hari, M.B.; Nair, S.; Kumar, V.A.; et al. Comparative molecular profiling of multidrug-resistant Pseudomonas aeruginosa identifies novel mutations in regional clinical isolates from South India. JAC-Antimicrob. Resist. 2024, 6, dlae001. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically engineered phages: A review of advances over the last decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef]
- Jo, S.J.; Kwon, J.; Kim, S.G.; Lee, S.-J. The biotechnological application of bacteriophages: What to do and where to go in the middle of the post-antibiotic era. Microorganisms 2023, 11, 2311. [Google Scholar] [CrossRef] [PubMed]
- Hesse, S.; Adhya, S. Phage therapy in the twenty-first century: Facing the decline of the antibiotic era; is it finally time for the age of the phage? Annu. Rev. Microbiol. 2019, 73, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Shende, R.K.; Hirpurkar, S.D.; Sannat, C.; Rawat, N.; Pandey, V. Isolation and characterization of bacteriophages with lytic activity against common bacterial pathogens. Vet. World 2017, 10, 973. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Breidt, F. Escherichia coli O157: H7 bacteriophage Φ241 isolated from an industrial cucumber fermentation at high acidity and salinity. Front. Microbiol. 2015, 6, 67. [Google Scholar] [CrossRef]
- Cockerill, F.R., III; Wikler, M.A.; Alder, J.; Dudley, M.N.; Eliopoulos, G.M.; Ferraro, M.J.; Hardy, D.K.; Hecht, D.W.; Hindler, J.A.; Patel, J.B. Approved Standard, 9th ed.; 2012 CLSI Document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Vipra, A.; Desai, S.N.; Junjappa, R.P.; Roy, P.; Poonacha, N.; Ravinder, P.; Sriram, B.; Padmanabhan, S. Determining the minimum inhibitory concentration of bacteriophages: Potential advantages. Adv. Microbiol. 2013, 3, 181–190. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, S.; Liu, Q.; Mai, G.; Yang, J.; Deng, D.; Zhang, B.; Liu, C.; Ma, Y. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front. Microbiol. 2018, 9, 1476. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. JoVE 2011, 47, 2437. [Google Scholar] [CrossRef]
- Lalitha, C.; Raman, T.; Rathore, S.S.; Ramar, M.; Munusamy, A.; Ramakrishnan, J. ASK2 bioactive compound inhibits MDR Klebsiella pneumoniae by antibiofilm activity, modulating macrophage cytokines and opsonophagocytosis. Front. Cell. Infect. Microbiol. 2017, 7, 346. [Google Scholar] [CrossRef]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 2010, 65, 1955–1958. [Google Scholar] [CrossRef]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.C.M.; de França, J.P.; Gaiba, S.; Aloise, A.C.; de Oliveira, A.F.; Moraes, A.A.d.F.S.; de França, L.P.; Ferreira, L.M. Development of experimental in vitro burn model. Acta Cir. Bras. 2014, 29 (Suppl. 2), 15–20. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Brister, J.R. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Wellcome Trust: London, UK, 2016; p. 84. [Google Scholar]
- Moghadam, M.T.; Amirmozafari, N.; Shariati, A.; Hallajzadeh, M.; Mirkalantari, S.; Khoshbayan, A.; Jazi, F.M. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect. Drug Resist. 2020, 13, 45–61. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and function of phage encoded depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef]
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic resistance increases with local temperature. Nat. Clim. Change 2018, 8, 510–514. [Google Scholar] [CrossRef]
- McArdle, C.D.; Lagan, K.M.; McDowell, D. A Effects of pH on the antibiotic resistance of bacteria recovered from diabetic foot ulcer fluid: An in vitro study. J. Am. Podiatr. Med. Assoc. 2018, 108, 6–11. [Google Scholar] [CrossRef]
- Bazbouz, M.B.; Tronci, G. Two-layer electrospun system enabling wound exudate management and visual infection response. Sensors 2019, 19, 991. [Google Scholar] [CrossRef]
- Spoering, A.L.; Lewis, K.I.M. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef]
- Kirker, K.R.; James, G.A. In vitro studies evaluating the effects of biofilms on wound--healing cells: A review. Apmis 2017, 125, 344–352. [Google Scholar] [CrossRef]
- Gonzalez, M.R.; Fleuchot, B.; Lauciello, L.; Jafari, P.; Applegate, L.A.; Raffoul, W.; Que, Y.-A.; Perron, K. Effect of human burn wound exudate on Pseudomonas aeruginosa virulence. Msphere 2016, 1, 10-1128. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).