Abstract
Chronic kidney disease (CKD) is a global public health issue that places an increasing burden on the healthcare systems of both the developed and developing countries. CKD is a progressive and irreversible condition, affecting approximately 10% of the population worldwide. Patients that have progressed to end-stage renal disease (ESRD) require expensive renal replacement therapy, i.e., dialysis or kidney transplantation. Current CKD therapy largely relies on the use of angiotensin-converting enzyme inhibitors (ACEis) and angiotensin receptor blockers (ARBs). However, these treatments by no means halt the progression of CKD to ESRD. Therefore, the development of new therapies is urgently needed. Antisense oligonucleotide (ASO) has recently attracted considerable interest as a drug development platform. Thus far, eight ASO-based drugs have been granted approval by the US Food and Drug Administration for the treatment of various diseases. Herein, we review the ASOs developed for the identification of CKD-relevant genes and/or the simultaneous development of the ASOs as potential therapeutics towards treating CKD.
1. Introduction
1.1. Antisense Oligonucleotide
Antisense oligonucleotides (ASOs) are one of the most classical synthetic therapeutic oligonucleotides that are able to modify gene expression. ASOs are typically designed to regulate gene expression by specifically binding to the pre-mRNA or mRNA of the gene via Watson–Crick base pairing [1,2,3,4]. Since the first report on the ASO-mediated inhibition of gene expression in the late 1970s by pioneering researchers Zamecnik and Stephenson [5,6,7], ASO technology has become a well-established platform for ASO-mediated RNA-targeting therapy [8]. So far, the US Food and Drug Administration (FDA) has granted approval for eight ASO drugs for clinical usage (Table 1 and Table S1) [9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25]. These successful clinical translations inspire both the academia and pharmaceutical industry to develop ASO-based drugs for the treatment of various diseases, either by the downregulation of disease-causing gene expression or rescuing the expression of essential but defective genes.

Table 1.
Antisense oligonucleotide (ASO)-based drugs approved by the FDA for clinical applications.
1.2. Chronic Kidney Disease
Chronic kidney disease (CKD) is a leading public health issue worldwide [26]. Approximately 850 million people (~10% adult population) are affected by CKD [27]. CKD accounted for 1.3% of years of life lost (YLL) in 2012 [28] and will become the fifth most common cause of YLL worldwide by 2040 [29]. In the US, CKD affects ~13% of the population [30], with more than 100,000 new patients starting on dialysis every year [31]. CKD is defined as the persistently decreased function of the kidney for more than 90 days [32]. Kidney dysfunction is shown by a glomerular filtration rate (GFR) of less than 60 mL/min/1.73 m2, or markers of kidney damage such as hematuria, albuminuria, and a variety of abnormalities detected by histology or imaging [32,33]. In CKD, the kidney undergoes a progressive and irreversible functional decline that can be classified into six stages (G1: normal, GFR ≥ 90 mL/min/1.73 m2; G2: 60–89 mL/min/1.73 m2; G3a: 45–59 mL/min/1.73 m2; G3b: 30–44 mL/min/1.73 m2; G4: 15–29 mL/min/1.73 m2; and G5: end-stage renal disease/kidney failure, <15 mL/min/1.73 m2). Patients who reach end-stage renal disease (ESRD) require kidney replacement therapy, i.e., dialysis or kidney transplantation, as their kidney is no longer able to maintain life in the long term [28]. However, less than half of patients needing kidney replacement therapy have access to treatment [34], as many governments and individuals are faced with the unaffordability of these costly therapies [35]. Moreover, kidney transplantation leads to a high risk of mortality as a result of unavoidable continuous immunosuppression (anti-rejection), which may cause infections and cancer development [36,37]. Therefore, it is imperative to delay or even prevent the progression of CKD from early stage to ESRD.
Although the causes of CKD vary geographically, diabetes accounts for 30–50% of CKD worldwide [28]. In developed countries, diabetes and hypertension are the leading causes of CKD [28]. Diabetic nephropathy is the main cause of ESRD in these countries and the burden of ESRD resulting from type 2 diabetes is projected to increase fourfold in decades to come [38,39], which is partly due to the increased prevalence of type 2 diabetes in young people [40]. In developing countries, CKD from glomerulonephritis and interstitial nephritis are more common as a result of the high prevalence of infections [41,42], such as streptococcal infections, acquired immunodeficiency syndrome (AIDS), schistosomiasis, leishmaniasis, hepatitis B, and hepatitis C. Notably, people affected by CKD are 5–10 times more likely to die prematurely due to the complications of CKD than ESRD [43]. This increased risk of death can be largely attributed to cardiovascular disease and cancer [44].
1.3. Cardiorenal Syndromes
Since the kidneys and heart have a bidirectional interorgan communication, dysfunction in one organ may cause dysfunction in the other organ, resulting in cardiorenal syndromes, a complex disorder of both the heart and kidneys [45]. Cardiorenal syndromes can be classified into five sub-types: type I [heart failure leading to acute kidney injury (AKI)], type II (chronic heart failure leading to CKD), type III (AKI leading to acute heart failure), type IV (CKD leading to heart failure), and type V (systemic condition such as diabetes mellitus leading to both renal and cardiac dysfunction) [45]. The common pathophysiological mechanisms in heart failure and CKD include dysfunction of the neurohormonal system (leading to the activation of the renin–angiotensin–aldosterone system), abnormal endothelial activation, reduced intestinal perfusion, and release of proinflammatory cytokines such as TNF-α, IL-1, and IL-6 [46]. These mechanisms simultaneously and sequentially contribute to cardiorenal syndromes, ultimately resulting in fibrosis and dysfunction in both organs [46].
2. Conventional Therapies and Their Limitations
Current standard of care for the treatment of CKD contains angiotensin-converting enzyme (ACE) inhibitors (ACEis) and angiotensin receptor (AR) blockers (ARBs) [47,48]. Drugs of these classes to some extent reduce the risk of kidney failure and major cardiovascular events [49,50]. The renoprotective effect of ACEis and ARBs is attributable to their ability to normalize glomerular hyperfiltration in kidneys [51,52]. However, both therapies slow but do not halt the progression of CKD from early stage to ESRD [53]. In order to improve their renal protection effect, combinational treatments including ACEi plus ARB, renin inhibitor plus ACEi, or renin inhibitor plus ARB were evaluated; however, the results were unsatisfactory in that unacceptable side effects such as hypotension, increased hyperkalemia, and AKI were observed, leading to the termination of the trials [54,55]. With the lack of clinically validated therapeutic targets (apart from ACE, AR, and renin) of CKD within or beyond the renin–angiotensin system, it is critically important to identify new targets for CKD in an attempt to develop novel therapeutics [53]. The targeted inhibition of gene expression by nucleic acid-based interventions can be a useful strategy for the identification and validation of potential CKD-relevant targets and the simultaneous or subsequent development of ASO-based therapy for CKD.
3. Antisense Oligonucleotide as Therapeutics
3.1. Mechanisms of Action
ASOs are short single-stranded chemically modified DNAs or RNAs that are about 15 to 30 nucleotides in length, complementary to their target pre-mRNA or mRNA. Upon binding specifically to their RNA targets and forming a target/ASO duplex, ASOs are capable of modulating gene expression through different mechanisms of action. These include: (1) induction of Ribonuclease H (RNase H)-mediated mRNA decay; (2) steric blockade that either modifies splicing by hindering the splicing factors from binding to pre-mRNA or represses gene expression by avoiding the 5′-capping of pre-mRNA or impeding the translational machinery from associating with mRNA (Figure 1) [2,56,57,58,59,60,61].
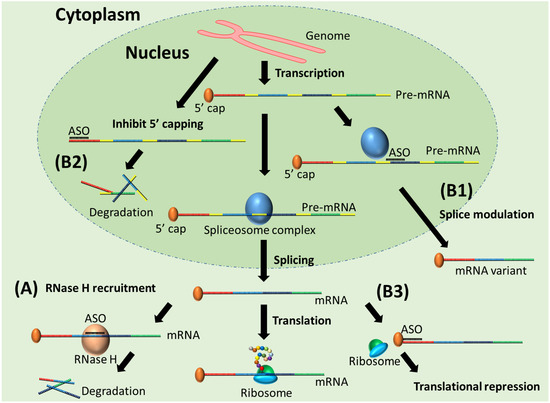
Figure 1.
Mechanisms of action of ASO. (A) RNase H recruitment by ASO/mRNA duplex and induced mRNA degradation; (B1) splice modulation, (B2) inhibition of 5′ capping, and (B3) translational arrest caused by steric-blocking ASOs.
RNase H-mediated mRNA degradation is the most used ASO mechanism for the purpose of gene knockdown, which allows for the exploration of gene function, identification and validation of potential disease-relevant genes, and therapeutic application by downregulating disease-causing gene expression [62]. The FDA has so far granted three RNase H-competent ASO drugs, fomivirsen (Vitravene®), mipomersen (Kynamro®), and inotersen (Tegsedi®) as therapies for cytomegalovirus (CMV) retinitis, familial hypercholesterolemia (FH), and hereditary transthyretin (TTR) amyloidosis, respectively [63]. Splice modulation by steric-blocking ASOs has been widely used to rescue the expression of essential but defective genes due to frameshifting mutations [64]. Splice modulation by exon skipping can restore the open reading frame by removing premature termination codons caused by mutations and restore the production of functional essential proteins. Since 2016, the FDA has approved five splice-modulating ASO drugs, eteplirsen (Exondys 51®), nusinersen (Spinraza®), golodirsen (Vyondys 53®), viltolarsen (Viltepso®), and casimersen (Amondys 45®) for the treatment of Duchenne muscular dystrophy (DMD) (eteplirsen, golodirsen, viltolarsen, casimersen) and spinal muscular atrophy (SMA) (nusinersen), respectively.
3.2. Chemical Modification and Rational Design of ASO
ASOs composed of natural deoxyribonucleotides or ribonucleotides are not suitable for research and/or therapeutic purposes as they are easily degraded by extracellular and intracellular nucleases, lacking target-binding affinity and specificity [63]. In order to resolve these problems, chemically modified nucleotides, i.e., nucleotide analogues, are used for constructing ASO sequences [65,66,67,68,69,70] (Figure 2). Each nucleotide comprises a sugar (either ribose or deoxyribose), a phosphate (internucleotide linkage), and a nitrogenous base, and these components can be chemically modified alone or combinedly. The earliest attempt to modify a nucleotide was focused on replacing the non-bridging oxygen atom of phosphate by other atoms or groups. Examples include phosphorothioate (PS), phosphorodithioate (PS2), methyl phosphonate (MP), boranophosphate (PB), and phosphonoacetate (PACE) [71,72,73,74,75] (Figure 2). PS modification (the oxygen is replaced by sulfur) enhances the nuclease stability of ASOs and prolongs their half-life in plasma. However, this advantage is compromised by increased toxicity. Mesyl phosphoramidate (MsPA) has attracted considerable attention recently as it shows significant advantages over PS in nuclease stability, binding affinity, and physiological safety [76,77,78]. Therefore, MsPA is considered a potential alternative to PS [76]. The sugar moiety of the nucleotide is the hotspot of modification. A number of nucleotide analogs with modified sugar have been discovered or invented, such as 2′-O-methyl (2′-OMe) [79,80], 2′-O-methoxyethyl (2′-MOE) [81], 2′-O, 4′-C-ethylenebridged nucleic acid (ENA) [82], locked nucleic acid (LNA) [83,84,85], threose nucleic acid (TNA) [86], 2′-amino (2′-NH2) [87], 2′-fluoro (2′-F) [88], 2′-fluroarabino nucleic acid (2′-FANA) [89], 1,5-anhydro hexitol nucleic acid (HNA) [90], cyclohexenyl nucleic acid (CeNA) [90], altritol nucleic acid (ANA) [90], and 2′-4′ constrained ethyl (cEt) [91] (Figure 2). Sugar modification may confer ASOs with improved nuclease resistance and enhanced hybridization affinity. Efforts have also been made on developing nucleotide analogues with a completely replaced backbone (i.e., sugar moiety plus phosphate moiety), such as peptide nucleic acid (PNA) [92] and phosphorodiamidate morpholino oligomer (PMO) [93] (Figure 2). A PNA monomer contains an uncharged N-(2-aminoethyl)-glycine, while a PMO monomer consists of a morpholine ring with a neutral phosphorodiamidate linkage. PNA and PMO display excellent target binding affinity and stability against cellular nuclease hydrolysis. Nucleobase modifications are not as commonly used as the above-described modifications on internucleotide linkage, sugar, or backbone in constructing ASO sequences [56].
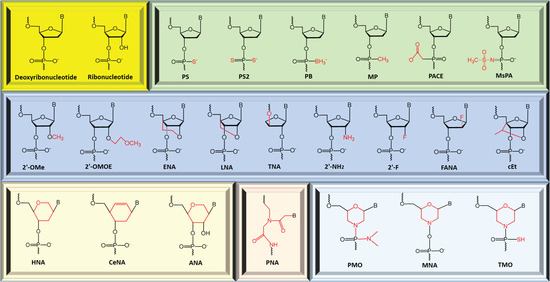
Figure 2.
Examples of modified nucleic acid monomer (nucleotide analogue). PS: phosphorothioate, PS2: phosphorodithioate, MP: methyl phosphonate, PB: boranophosphate, PACE: phosphonoacetate, MsPA: mesyl phosphoramidate, 2′-OMe: 2′-O-methyl, 2′-MOE: 2′-O-methoxyethyl, ENA: 2′-O, 4′-C-ethylenebridged nucleic acid, LNA: locked nucleic acid, TNA: threose nucleic acid, 2′-NH2: 2′-amino, 2′-F: 2′-fluoro, 2′-FANA: 2′-fluroarabino nucleic acid, HNA: 1,5-anhydro hexitol nucleic acid, CeNA: cyclohexenyl nucleic acid, ANA: altritol nucleic acid, cEt: 2′-4′ constrained ethyl, PNA: peptide nucleic acid, PMO: phosphorodiamidate morpholino oligomer, MNA: morpholino nucleic acid, and TMO: thiophosphoramidate morpholino oligomer.
An ASO can be synthesized as a uniformly modified sequence, a gapmer, or a mixmer (Figure 3 and Figure S1). Uniform modification and mixmer-design confer ASO maximized improvement in terms of nuclease stability and binding affinity. However, most of the modifications (except for the above-mentioned modified linkages PS, PS2, PB, PACE, MsPA, excluding MP) are unable to induce target RNA degradation as these analogues do not support RNase H-mediated cleavage. Therefore, uniformly modified ASOs are solely used as a steric blocker to induce splice switching or translational repression. In order to retain the ability of ASOs in recruiting RNase H and improve their nuclease stability at the same time, gapmer design emerges as the perfect solution [94,95,96,97]. A gapmer-like ASO usually consists of an unmodified or PS-modified central sequence (~10 deoxyribonucleotides) and sugar moiety-modified flanking sequences (~5 nucleotide analogues) on both its sides (Figure 3 and Figure S1). The central DNA region ensures that the ASO is capable of inducing RNase H-dependent target hydrolysis, while the flanking modified sequences protect the ASO from nuclease attack and enhance binding affinity. Most recently, Anderson et al. reported that combination of PS and MsPA linkages could further improve the therapeutic index and duration of effect of gapmer-like ASOs [78]. Most of the nucleotide analogues can be used to synthesize chimeric ASOs (i.e., gapmer and mixmer) that contain more than one chemistry in an attempt to optimize the stability and efficacy of ASOs. However, it has been challenging to develop a flexible and robust synthetic route that allows for the generation of chimeric PMO sequencing with other analogues. To overcome this, Veedu et al. developed an analogue of PMO called morpholino nucleic acid (MNA), allowing the synthesis of chimeric ASO with both morpholine ring-containing nucleotide and 2′-OMe nucleotides, providing a promising solution for the incompatibility of PMO [98]. More recently, Caruthers et al. reported the synthesis of a new analogue called thiophosphoramidate morpholino (TMO), which allows the easy incorporation of morpholino–PS moieties and other sugar modifications [99]. To date, PS, 2′-MOE, and PMO are the only three chemistries used in FDA-approved ASO drugs. Specifically, fomivirsen is a 21 mer DNA sequence uniformly modified by PS [9,10]; both mipomersen and inotersen are 20 mer 5-10-5 2′-MOE-PS gapmers (5-10-5 MOEPS-DNAPS-MOEPS design) [11,12,21,22], i.e., they contain a central 10 mer DNA-PS sequence and two 5 mer flanking 2′-MOE-PS sequences; nusinersen is an 18 mer ASO uniformly modified by 2′-MOE-PS [17,18,19,20]; and the four DMD-targeting fully PMO-modified drugs, eteplirsen, golodirsen, viltolarsen, and casimersen have 30, 25, 21, and 22 monomers, respectively [13,14,15,16,23,24,25].
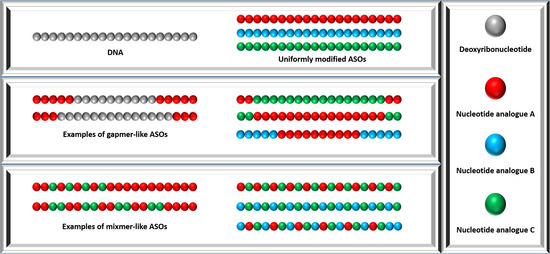
Figure 3.
Design of chemically modified ASOs including uniformly modified ASO design, gapmer design, and mixmer design.
3.3. ASO, siRNA, and miRNA
ASO and small interfering RNA (siRNA) are the two most widely used nucleic acid-based strategies for the transient silencing of gene expression [100]. Different from ASO, siRNA has two strands and degrades target mRNA by RNA-induced silencing complex (RISC) instead of RNase H [101,102,103]. Since unmodified RNAs possess a higher potency than oligodeoxynucleotides, it is relatively easier to obtain a potent siRNA than an ASO. Therefore, siRNA is considered a more preferable option for in vitro studies [101]. Although lower doses of therapeutic siRNAs are sufficient to cause target gene knockdown, and they exhibit a longer duration of activity as compared to common ASOs, off-target effects of siRNAs (such as microRNA-like off-target effects and activation of Toll-like receptors) can lead to toxicities which compromise their therapeutic benefits [104]. Some of the off-target effects could be alleviated by pinpointing chemical modification (e.g., the seed region of siRNA) [105]. However, ASO is the better choice to be developed as nucleic acid-based therapeutics for four reasons: (1) lower cost of production as ASOs are single stranded, (2) in vivo delivery of ASOs is easier than siRNA (ASO delivery does not need a vector while siRNA delivery needs a carrier) [101], (3) ASOs can not only silence gene expression, but also restore gene expression which has been recognized as the most effective strategy for the treatment of DMD and SMA, (4) novel chemical modification (e.g., MsPA) and its rational positioning in gapmer-like ASOs lead to the advent of very long-acting next generation antisense oligomers [78].
MicroRNAs (miRNAs) are short noncoding endogenous RNAs of 18–22 mer in length that play important roles in the up- or down-regulation of genes [106]. miRNA-based therapeutic oligonucleotides can be developed as miRNA mimics or anti-miRNAs (ASOs). miRNA mimics imitate miRNA functions, while anti-miRNAs bind to miRNAs complementarily and deactivate their functions through RNase H-mediated degradation or the steric blockage mechanism [107,108]. Like the chemically modified ASOs targeting mRNAs/pre-mRNAs, anti-miRNAs are usually composed of nucleotide analogues instead of unmodified nucleotides to obtain improved nuclease stability and target miRNA-binding affinity. For example, miRNA-92a is a potential therapeutic target of cardiovascular diseases with CKD as it mediates endothelial dysfunction in CKD [109]. Hinkel et al. developed an LNA-modified ASO as an anti-miRNA-92a (named LNA-92a) [110], which efficiently inhibited miRNA-92a leading to protection against ischemia-reperfusion injury in a pig model. In addition, long non-coding RNAs (lncRNAs) regulate the effects of miRNAs on mRNA expression [111]. Some of the lncRNAs have emerged as potential prognostic biomarkers for CKD progression, such as lncRNAs HCP5, and NOP14-AS1 [112]. Development of ASOs targeting these lncRNAs could facilitate research aiming at elucidating their roles in CKD and developing ASO-based therapeutic strategies.
4. Antisense Oligonucleotides Targeting Chronic Kidney Disease
ASO-mediated gene knockdown by specifically reducing the mRNA production of genes enables the identification and/or verification of disease-relevant genes in CKD, leading to the expansion of knowledge regarding the molecular basis underlying the disease, thereby bringing new hope to identifying promising target genes for CKD therapy aimed at delaying or halting the progressive decline of kidney function. Furthermore, upon recognition of genes as potential therapeutic targets for CKD, the identified ASO-based gene inhibitors can directly serve as lead compounds, which speeds up the drug development process and bypasses the potential difficulties that development of small molecule-based inhibitors may face, such as the undruggability of target proteins and the toxicity of small molecules.
In fact, uniformly PS-modified ASOs and 2′-MOE-PS gapmer-like ASOs have been developed by different groups to target genes involved in the progression of CKD by downregulating the expression of those genes through the RNase H mechanism. In this section, we review the ASO-based research on genes that are relevant to CKD, including THBS1 (also known as TSP1) that encodes thrombospondin-1 (TSP1), CCN2 (also known as CTGF) encoding connective tissue growth factor (CTGF), KRAS encoding Kirsten rat sarcoma viral oncogene homolog (KRAS), MTOR encoding mammalian target of rapamycin (mTOR), AGT encoding angiotensinogen (AGT), and APOL1 that encodes apolipoprotein L1 (APOL1). These studies provide new insights into the molecular pathogenesis of CKD and explore the therapeutic potential of ASO-based gene inhibitors for the targeted therapy of CKD. In vitro screening of ASO candidates and subsequent in vivo investigations in these studies are shown in Table 2 and Table 3, respectively.

Table 2.
In vitro screening of ASOs and the identified best-performing candidates for subsequent in vitro and/or in vivo studies. Purple asterisks “*” in ASO sequences represent PS modification, red color represents 2′-MOE modification, blue color represents 2′-4′ constrained ethyl modification, black color represents deoxyribonucleotide. 2′-MOE: 2′-O-methoxyethyl, PS: Phosphorothioate internucleotide linkage, PO: Phosphodiester internucleotide linkage.

Table 3.
Representative results of in vivo studies of ASO-mediated silencing of CKD-related genes.
4.1. Thrombospondin-1 (TSP1)
Renal fibrosis, defined as the pathological accumulation of extracellular matrix, is the common hallmark of CKD [119]. Transforming growth factor-β (TGF-β), a profibrotic cytokine, plays a major role in experimental renal disease when TGF-β is overexpressed in the anti-Thy1 model (induced mesangial proliferative glomerulonephritis in rats) [120,121], and accumulation of mesangial cell matrix and interstitial fibrosis could be identified in transgenic mice expressing active TGF-β1 [122]. Furthermore, in human CKD, the upregulation of TGF-β is correlated with excess extracellular matrix, providing evidence that TGF-β plays a vital role in mediating fibrosis [123]. To this end, Akagi et al. directly blocked TGF-β action in the anti-Thy1 model by the administration of TGF-β-targeting PS-modified DNA ASOs, which achieved markedly reduced extracellular matrix accumulation [124]. However, TGF-β is not a suitable therapeutic target as it is a pleiotropic cytokine that exhibits other essential biofunctions in mammals, and TGF-β knockout mice survive for only a few weeks after birth [125,126,127].
Thrombospondin-1 (TSP1) is a major activator of TGF-β1 [128,129,130,131]. Nevertheless, unlike the TGF-β null mice, TSP1 knockout mice do not die prematurely and are healthy [130,131]. De novo TSP1 expression in mesangial cells colocalizes with the upregulation of TGF-β1 in different experimental kidney disease models [132], including the anti-Thy1 model [121,133]. In order to investigate the role of TSP1 as a TGF-β activator in the development of renal fibrosis, Daniel et al. screened out two PS-modified DNA ASO sequences (an 18 mer ASO and a 15 mer ASO) from 11 candidates that were designed to inhibit TSP1 expression [113]. ASOs were selectively transferred to the glomeruli of Sprague-Dawley (SD) rats with anti-Thy1 antibody-induced mesangial proliferative glomerulonephritis through renal artery perfusion followed by electroporation. Six days after treatment, TSP1-specific ASOs achieved the efficient reduction of TSP1 expression by more than 60%, and the inhibition of active TGF-β secretion by 50%, while not affecting the total expression level of TGF-β [113]. The ASO treatment also led to an evident reduction in the glomerular number of nucleic positive for the phospho-Smad2/3 (a TGF-β-signaling molecule that is known as a marker of TGF-β activation), indicating a markedly decreased glomerular TGF-β activity, which was associated with a marked reduction in mesangial cell activation [113]. Furthermore, TSP1-targeting ASO therapy inhibited the accumulation of glomerular extracellular matrix so that extra-domain A of fibronectin was markedly reduced by nearly 96%, as well as the reduced accumulation of other extracellular matrix proteins such as collagen I and collagen IV [113]. This study reveals that TSP1 is a tightly regulated activator of TGF-β in the anti-Thy1 model, and is responsible for the accumulation of glomerular extracellular matrix by activating TGF-β. Moreover, the data of the study suggests that the ASO-based TSP1 inhibitor may be a feasible therapeutic for fibrotic renal disease.
4.2. Connective Tissue Growth Factor (CTGF)
Over 30% of ESRD is caused by diabetic nephropathy. Glomerulosclerosis, the pathological hallmark of diabetic nephropathy, is characterized by the extracellular matrix accumulation of mesangial cells and tubulointerstitial fibrosis [134]. Both in vitro and in vivo studies have established that TGF-β contributes to glomerulosclerosis and that overexpression of TGF-β is associated with fibrosis and scarring in response to renal injury in diabetes [135,136,137,138,139]. However, as mentioned in the previous section, TGF-β is not a suitable target for drug development due to its multifunctionality. CTGF, a prosclerotic cytokine overexpressed during diabetes and acting downstream of TGF-β [140], directly contributes to the accumulation of extracellular matrix and tubulointerstitial fibrosis in diabetic nephropathy [140,141,142]. Okada et al. downregulated the expression of CTGF in tubular epithelium by the intravenous administration of a CTGF-targeting, PS-modified 18 mer DNA ASO in mice with subtotal nephrectomy (SNx) [143]. They found that decreased expression of CTGF caused by the ASO is associated with attenuated interstitial fibrosis resulting from the downregulated expression of genes involved in the expansion of the glomerular extracellular matrix, demonstrating that CTGF is a direct and significant contributor of TGF-β-dependent renal fibrogenesis [143]. This study suggested that the development of an ASO-based CTGF inhibitor may be a promising strategy for antifibrotic therapy in TGF-β-dependent CKD such as diabetic nephropathy.
Later, Guha et al. investigated the role of CTGF in the progression of diabetic nephropathy by administrating a CTGF-specific 20 mer ASO (either 4-12-4 MOEPO-DNAPS-MOEPO or 4-12-4 MOEPS-DNAPS-MOEPS gapmer) to nephropathic mice with streptozotocin-induced type 1 diabetes and db/db mice (mice with type 2 diabetes) with a dosage of 20 mg/kg (twice weekly) for 16 weeks and 8 weeks, respectively [114]. The CTGF-targeting ASO-inhibited hyperglycemia induced overexpression of CTGF in both diabetic mouse models. In the mice with type 1 diabetes, ASO treatment inhibited a variety of indices of renal disease, for instance, the development of renal hypertrophy was significantly attenuated in that the diabetes-induced increase of kidney weight was reduced by 32%, increases of serum creatinine and urinary albumin which are pathological features of diabetic nephropathy were reduced by 32% and 52%, respectively, and the diabetes-induced expansion of the mesangial matrix was attenuated by 43%, which was associated with the reduced synthesis of collagen 1, fibronectin, and TGF-β1 [114]. Furthermore, ASO treatment achieved the inhibition of profibrotic p38 mitogen-activated protein kinase (MAPK) and its downstream target transcription factor cAMP-response element binding protein (CREB) so that the activation of p38 MAPK and CREB was attenuated by 54% and 74%, respectively, indicating that the progression of diabetic nephropathy may be inhibited [114]. In addition, in the db/db mice, CTGF-targeting ASO reduced serum creatinine, urinary total protein, and urinary albumin by 37%, 41%, and 48%, respectively [114]. This study provides sound scientific evidence that the specific knockdown of CTGF by gapmer-like ASO holds significant promise as a potential therapy for diabetic nephropathy.
4.3. Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS)
Tubulointerstitial fibrosis, characterized by the excessive deposition of extracellular matrix resulting from an increased number of activated interstitial myofibroblasts, is a key determinant of progressive CKD [144]. RAS proteins, termed small guanosine triphosphatases (GTPases), play essential roles in the regulation of cell survival, proliferation, and differentiation by acting as signal transduction molecules in various extracellular pathways [145]. Bechtel et al. demonstrated that the activation of RAS is directly associated with renal fibrogenesis [146]. Therefore, RAS proteins may be potential therapeutic targets for the renal fibrosis of CKD [147,148,149,150,151,152,153,154]. Sharpe et al. demonstrated that KRAS is the predominant isoform of RAS expressed in human renal fibroblasts and that PS or 2′-MOE-PS-modified ASO-induced KRAS knockdown significantly suppresses the proliferation of fibroblasts [155,156]. Later, in order to investigate the profibrotic role of KRAS in CKD, the same group silenced the KRAS expression in rats with unilateral ureteric obstruction (unilateral ureteric obstruction is considered a model of renal fibrosis and CKD [157]) by the subcutaneous administration of KRAS-specific 20 mer gapmer-like ASOs (ISIS 104440 or ISIS 104419, 5-10-5 MOEPS-DNAPS-MOEPS design) on alternate days (for six days) at a dosage of 12.5 mg/kg [115]. Treatment of KRAS-specific ASOs significantly reduced the level of KRAS mRNA by 61% (ISIS 104440) and 97% (ISIS 104419) compared to their correspondent scrambled ASOs (negative control), which was associated with reduced renal fibrosis (fibrosis score was reduced to 17% by ISIS 104440 and 20.3% by ISIS 104419) and collagen deposition (collagen deposition score was reduced to 18.4% by ISIS 104440 and 17% by ISIS 104419) [115]. Furthermore, the upregulation of α-smooth muscle actin (α-SMA, a marker of myofibroblast activity) induced by obstructive nephropathy was inhibited by the KRAS-targeting ASOs so that the α-SMA expression was reduced from 53% (negative control group) to 3.9% and 20% by ISIS 104440 and ISIS 104419, respectively, indicating that an ASO-mediated KRAS knockdown could prevent the onset of fibrosis in rat models of unilateral ureteric obstruction [115]. This study suggests that ASO-induced KRAS inhibition could be a novel antifibrotic strategy for CKD as ASO treatment markedly inhibited renal fibrosis.
It is worth mentioning that Ross et al. recently evaluated another highly potent KRAS-specific ASO candidate (AZD4785 or ISIS 651987) for its anti-tumour effects in vitro and in vivo [158]. We recommend researchers compare the efficacy between AZD4785 and ISIS 104440/ISIS 104419 to identify the best-performing lead candidate for further anti-CKD therapeutic development.
4.4. Mammalian Target of Rapamycin (mTOR)
Autosomal dominant polycystic kidney disease (PKD), caused by a mutation in the PKD1 or PKD2 gene, with 50% of patients developing CKD, accounts for ~5–10% of ESRD in the US requiring dialysis or renal transplantation [159]. The mTOR signaling pathway plays a role in the regulation of cell growth and proliferation [160]. Specifically, mTOR complex 1 (mTORC1) controls protein synthesis and cell proliferation, and the hyperactivation of the mTORC1 signal is a feature of PKD [161,162,163,164]. mTOR complex 2 (mTORC2) modulates cell survival and arrangement of actin cytoskeleton by phosphorylating AGC kinases such as the pro-survival kinase pAktSer473 [165,166,167]. Activation of mTORC2 is upregulated in PKD [162,163,164,165,166,167,168,169]. Sirolimus, an mTORC1 inhibitor, has been demonstrated to have a therapeutic effect on mice with PKD resulting from Pkd1 inactivation via the reduction of cyst growth and preservation of renal function [162]. However, sirolimus had no effect on renal function in Pkd2WS25/− mice (a model of human autosomal dominant PKD resulting from mutation in the Pkd2 gene) [170], and a clinical study showed that sirolimus treatment did not halt the growth of polycystic kidneys in humans [171]. One possible reason of the inefficacy is that sirolimus directly inhibits mTORC1 but not mTORC2, therefore, it is unable to inhibit mTORC2-dependent Akt-induced proliferation [172].
In order to determine the therapeutic effect of the combined inhibition of mTORC1 and mTORC2 on PKD, Ravichandran et al. developed an mTOR-specific 20 mer gapmer-like ASO (5-10-5 MOEPS-DNAPS-MOEPS design) through the screening of ∼150 ASO candidates [116], as mTOR exists in both mTORC1 and mTORC2 [173]. ASOs were administered into Pkd2WS25/− mice via intraperitoneal injections at a sequential two-stage dosage of 100 mg/kg/week (first 4 weeks) and 50 mg/kg/week (the remaining 8 weeks) [116]. Treatment of mTOR ASO led to a significant reduction in the expression levels of mTOR, pS6 (a marker of mTORC1 signaling), and pAktSer473 [116], which was associated with the reduced ratio of two kidneys/total body weight from 2.4% (control ASO group) to 1.5% (mTOR ASO group), significantly decreased cyst volume density from 34.1% (control ASO group) to 15.1% (mTOR ASO group), and significantly reduced blood urea nitrogen from 43.4 mg/dL (control ASO group) to 29 mg/dL (mTOR ASO group) [116], indicating an ASO-induced melioration of PKD and normalization of renal function. Furthermore, mTOR ASO treatment significantly inhibited both the proliferation and apoptosis of tubular epithelial cells (proliferation and apoptosis of epithelial cells lining the renal tubular plays a key role in cyst growth [174]) so that the number of proliferating cell nuclear antigen positive cells was reduced from 1.9 per cyst (control ASO group) to 0.8 per cyst (mTOR ASO group), and the number of apoptotic cells was reduced from 3.6 per non-cystic tubule (control ASO group) to 1.2 per non-cystic tubule (mTOR ASO group) [116], suggesting that ASO therapy could inhibit cyst growth. This study demonstrated that the combined inhibition of both mTORC1 and mTORC2 holds therapeutic potential for autosomal dominant PKD [116], and an ASO-based mTOR inhibitor can be a promising approach for treating the disease owing to its capability of combined mTORC1/2 knockdown.
4.5. Angiotensinogen (AGT)
Enlargement of renal cysts in autosomal dominant PKD is associated with the activation of the renin–angiotensin system and the resultant production of proinflammatory and profibrotic angiotensin II [175,176,177], which contributes to cystogenesis by the induction of cellular proliferation, inflammation, and fibrosis [175,178]. The renin–angiotensin system also plays an important role in type IV cardiorenal syndrome and CKD [179]. Angiotensin II induces the differentiation of renal fibroblasts into myofibroblasts and stimulates the expression and activation of TGF-β [180,181]. Angiotensin II leads to renal damage by increasing the expression of proinflammatory cytokines and chemokines and renal leukocyte infiltration [182]. Despite the importance of the renin–angiotensin system in the pathogenesis of PKD, the single or combined use of renin–angiotensin system inhibitors, including ACEis, ARBs, and renin inhibitor, have limited efficacy and induce side effects [54,55]. Identification of other targets within the renin–angiotensin system may result in the development of more efficient therapeutics.
Angiotensinogen (AGT) is a substrate of the peptidase renin in the renin–angiotensin system, and AGT is cleaved by renin forming angiotensin I, which is then converted to angiotensin II by ACE [181]. In order to investigate the potential therapeutic effects of direct AGT inhibition on PKD, Ravichandran et al. developed an AGT-specific 20 mer gapmer-like ASO (5-10-5 MOEPS-DNAPS-MOEPS design) through the screening of ∼150 ASO candidates [117]. The administration of ASO was performed by intraperitoneal injections into Pkd2WS25/− mice at a sequential two-stage dosage of 100 mg/kg/week (first 4 weeks) and 50 mg/kg/week (the remaining 8 weeks) [117]. AGT-specific ASO treatment significantly reduced the AGT expression, which was associated with a reduced two kidney/total body weight ratio (AGT ASO group: 1.5%, control ASO group: 2.4%), decreased cyst volume density (AGT ASO group: 22%, control ASO group: 34.1%) and blood urea nitrogen (AGT ASO group: 34 mg/dL, control ASO group: 47 mg/dL), indicating that AGT-specific ASO treatment could lead to decreased PKD and normalization of renal function [117]. Furthermore, significant decreases in proinflammatory cytokines including C-X-C motif chemokine ligand 1 (CXCL1), interleukin 12 (IL-12), and the profibrotic TGF-β were observed in the AGT ASO treatment groups (CXCL1: 0.6 pg/mg, IL-12: 8.8 pg/mg, TGF-β: 32 pg/mg) in contrast to the control ASO treatment groups (CXCL1: 3.4 pg/mg, IL-12: 37.3 pg/mg, TGF-β: 102 pg/mg), indicating that AGT-specific ASO treatment could lead to decreased proinflammatory and profibrotic molecules [117]. Although further investigation is required to elucidate the mechanism underlying the ASO-induced inhibition on cyst growth, chronic AGT inhibition by ASO may be a possible therapeutic strategy for autosomal dominant PKD in the future.
4.6. Apolipoprotein L1 (APOL1)
APOL1 is a newly evolved gene that is only present in a few primates such as humans, baboons, and gorillas [183,184]. The APOL1 protein functions as the trypanolytic factor in serum that lyses trypanosomes against African trypanosomiasis (sleeping sickness), a disease endemic to Africa [185,186,187]. One of the trypanosome species, Trypanosoma brucei rhodesiense, has evolved to resist the wild type APOL1 (G0), while the G1 and G2 mutants of APOL1 (commonly found in populations of African ancestry), discovered in 2010, overcome the resistance of Trypanosoma brucei rhodesiense [188]. However, mutant APOL1 also leads to toxic gain of function when overexpressed so that [189,190], compared to the wild type APOL1 (G0), the G1/G2 mutants are associated with an increased risk of CKD by 7- to 30-fold [188,191,192,193]. Furthermore, these mutants accelerate the GFR decline, and thus the progression of CKD [192,194]. In addition, G1/G2 mutants are strongly associated with various forms of nondiabetic nephropathy such as focal segmental glomerulosclerosis (FSGS) and interferon (IFN) therapy-related collapsing glomerulopathy [188,189,192]. As APOL1 is not essential for kidney development and function [183,195,196,197], reducing the expression level of APOL1 therapeutically will not result in any harmful effects other than increased susceptibility to African sleeping sickness in specific geographical regions [118]. Therefore, in an attempt to study APOL1 systemically and achieve proof of concept for APOL1 inhibition by antisense oligomer, Aghajan et al. established a transgenic mouse model for APOL1-related CKD (C57BL/6 mice with human APOL1 G1 mutant gene were challenged by IFN-γ leading to induced proteinuria in mice), developed a APOL1 specific, 2′-MOE or 2′-4′ constrained ethyl (cEt)-modified 16 mer gapmer ASO on a PS backbone (IONIS-APOL1Rx) through the screening of over 4000 ASO candidates, and treated APOL1 G1 transgenic mice with IONIS-APOL1Rx at a weekly dose of 50 mg/kg for four weeks prior to IFN-γ challenge [118]. ASO treatment led to a significant decrease in the APOL1 mRNA levels in both the kidney (by ~50%–60%) and liver (by 95%), which completely prevented the occurrence of IFN-γ-triggered proteinuria, indicating that the CKD-relevant cell types (podocytes, endothelial cells, and mesangial cells that constitute the renal filtration barrier) are sensitive to ASO treatment [118]. It was also found that administration of IONIS-APOL1Rx at a weekly dosage as low as 6.25 mg/kg led to the significant inhibition of induced proteinuria, demonstrating the potency of ASO therapy and the renoprotective effect that it provided [118]. Although further study is definitely required to reveal the pathogenesis of the toxic gain of function resulting from mutant APOL1, IONIS-APOL1Rx holds promise to be developed as an efficient anti-CKD therapeutic option for patients with APOL1 nephropathies.
5. Potential Problems of ASO-Based CKD Therapy and Possible Solutions
The efficient delivery of therapeutic ASOs to their target tissues remains a major challenge. Although all FDA-approved ASO drugs are administered directly by injection, intense research efforts have yielded different strategies aimed at improving the in vivo delivery of ASOs, such as nanocarriers, viral delivery, nanoparticles, antibodies, and aptamers. The advancement in the delivery methods of therapeutic oligonucleotides has been discussed in a number of recent reviews [198,199,200,201,202].
The ASO-mediated knockdown of CKD relevant gene expression may lead to severe adverse effects due to the multifunctionality of target genes. For example, although TGF-β inhibition could suppress progression to glomerulosclerosis [123], TGF-β knockout could be deadly due to its essential roles in multiple developmental processes [124,125,126]. Possible solutions include:
- (1)
- Exploration of target genes that are located upstream or downstream of TGF-β pathway, such as TSP1 (upstream) [113] and CTGF (downstream) [114].
- (2)
- Exploration of target genes that are newly evolved or less conserved, such as APOL1 [118], so that inhibition of such genes is probably less risky compared with the genes that are functionally conserved.
- (3)
- Identification of causative genes for diabetes and hypertension as these diseases are the leading causes of CKD; ASO-based targeted therapies may prevent the induction of CKD [8,26,28,203].
- (4)
- Investigation of AKI-related biomarkers such as neutrophil gelatinase-associated lipocalin (NGAL) and kidney injury molecule-1 (KIM-1) [204,205], since these biomarkers could be potential therapeutic targets that could be involved in the AKI-to-CKD transition.
Off-target effects of ASO that are either sequence dependent or independent may result in in vivo toxicity. Possible solutions include: (1) extending the length of ASO from 15 mer to ~25–30 mer, thus increasing its specificity and reducing potential hybridization between ASO and non-target RNAs; (2) employment of nucleotide analogues of improved safety profile, such as PMO to reduce the sequence independent toxicity of ASO. One example of this solution is the rejection of drisapersen (2′-OMe-PS modified ASO, BioMarin Pharmaceutical) due to life-threatening toxicity and the approval of eteplirsen (PMO modified ASO, Sarepta Therapeutics) by the FDA in 2016 (both drisapersen and eteplirsen are DMD mRNA exon-51-targeting drugs) [56].
6. Conclusions
Patients with CKD progressed to ESRD require expensive dialysis or kidney transplantation; however, current CKD therapy relying largely on the use of renin–angiotensin system inhibitors, such as ACEis and ARBs, does not halt the progression of CKD. Therefore, the development of new therapies with an improved potency and safety profile is urgently needed. Chemically modified ASOs have been used in the functional study of genes involved in the pathogenesis of CKD and the subsequent or coinstantaneous development of ASO-based inhibitors of the target genes. Compared with conventional small molecule-based protein inhibitors, ASOs possess an advantage due to their capability of inducing direct targeted mRNA degradation, bypassing potential obstacles facing “undruggable” proteins. As a result, a few ASO-based inhibitors of genes responsible for the pathogenesis of CKD have been developed. It is recommended that the antifibrotic effects of these ASOs in the heart should also be investigated since cardiac and renal dysfunction share common mechanisms. Furthermore, efforts have been made in the identification of novel therapeutic targets (other than the ACE and AR) within or beyond the renin–angiotensin system. Given the convenience and flexibility of developing an ASO-based inhibitor of a specific gene, there is no doubt that more CKD-relevant genes will be identified as potential therapeutic targets, which may lead to the development and approval of antisense anti-CKD drugs in the future.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/kidneydial2010004/s1, Table S1: Results of clinical investigations of FDA-approved ASOs. Figure S1: The relationship between ASO design and the mechanism of action of ASOs.
Author Contributions
Conceptualization, S.C. and T.Z.; writing—original draft preparation, Y.L. and S.C.; writing—review and editing, Y.L., Y.T., R.Z., T.W., N.N., T.Z., R.N.V., S.C.; supervision, S.C.; funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a Ph.D. startups grant provided by the School of Food and Biological Engineering, Henan University of Animal Husbandry and Economy.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alama, A.; Barbieri, F.; Cagnoli, M.; Schettini, G. Antisense oligonucleotides as therapeutic agents. Pharmacol. Res. 1997, 36, 171–178. [Google Scholar] [CrossRef]
- Dias, N.; Stein, C.A. Antisense oligonucleotides: Basic concepts and mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [Google Scholar]
- Crooke, S.T.; Baker, B.F.; Crooke, R.M.; Liang, X.H. Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 2021, 20, 427–453. [Google Scholar] [CrossRef]
- Chan, J.H.; Lim, S.; Wong, W.S. Antisense oligonucleotides: From design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 533–540. [Google Scholar] [CrossRef]
- Paterson, B.M.; Roberts, B.E.; Kuff, E.F. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proc. Natl. Acad. Sci. USA 1977, 74, 4370–4374. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288. [Google Scholar] [CrossRef]
- Chen, S.; Sbuh, N.; Veedu, R.N. Antisense oligonucleotides as potential therapeutics for type 2 diabetes. Nucleic Acid Ther. 2021, 31, 39–57. [Google Scholar] [CrossRef]
- Roehr, B. Fomivirsen approved for CMV retinitis. J. Int. Assoc. Physicians AIDS Care 1998, 4, 14–16. [Google Scholar]
- De Smet, M.D.; Meenken, C.J.; van den Horn, G.J. Fomivirsen—A phosphorothioate oligonucleotide for the treatment of CMV retinitis. Ocul. Immunol. Inflamm. 1999, 7, 189–198. [Google Scholar] [CrossRef]
- Hair, P.; Cameron, F.; McKeage, K. Mipomersen sodium: First global approval. Drugs 2013, 73, 487–493. [Google Scholar] [CrossRef]
- Wong, E.; Goldberg, T. Mipomersen (kynamro): A novel antisense oligonucleotide inhibitor for the management of homozygous familial hypercholesterolemia. Pharmacol. Ther. 2014, 39, 119–122. [Google Scholar]
- Syed, Y.Y. Eteplirsen: First global approval. Drugs 2016, 76, 1699–1704. [Google Scholar] [CrossRef]
- Lim, K.R.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 2017, 11, 533–545. [Google Scholar] [CrossRef]
- Baker, D.E. Eteplirsen. Hosp. Pharm. 2017, 52, 302–305. [Google Scholar] [CrossRef]
- Charleston, J.S.; Schnell, F.J.; Dworzak, J.; Donoghue, C.; Lewis, S.; Chen, L.; Young, D.; Milici, A.; Voss, J.; DeAlwis, U.; et al. Eteplirsen treatment for Duchenne muscular dystrophy: Exon skipping and dystrophin production. Neurology 2018, 90, e2146–e2154. [Google Scholar] [CrossRef]
- Hoy, S.M. Nusinersen: First global approval. Drugs 2017, 77, 473–479. [Google Scholar] [CrossRef]
- Corey, D.R. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 2017, 20, 497–499. [Google Scholar] [CrossRef]
- Goodkey, K.; Aslesh, T.; Maruyama, R.; Yokota, T. Nusinersen in the treatment of spinal muscular atrophy. Methods Mol. Biol. 2018, 1828, 69–76. [Google Scholar]
- Neil, E.E.; Bisaccia, E.K. Nusinersen: A novel antisense oligonucleotide for the treatment of spinal muscular atrophy. J. Pediatr. Pharmacol. Ther. 2019, 24, 194–203. [Google Scholar] [CrossRef]
- Keam, S.J. Inotersen: First global approval. Drugs 2018, 78, 1371–1376. [Google Scholar] [CrossRef]
- Gales, L. Tegsedi (inotersen): An antisense oligonucleotide approved for the treatment of adult patients with hereditary transthyretin amyloidosis. Pharmaceuticals 2019, 12, 78. [Google Scholar] [CrossRef]
- Heo, Y.A. Golodirsen: First approval. Drugs 2020, 80, 329–333. [Google Scholar] [CrossRef]
- Dhillon, S. Viltolarsen: First approval. Drugs 2020, 80, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.C.; Zhang, L.X. Prevalence and disease burden of chronic kidney disease. Adv. Exp. Med. Biol. 2019, 1165, 3–15. [Google Scholar] [PubMed]
- Li, P.K.T.; Garcia-Garcia, G.; Lui, S.F.; Andreoli, S.; Fung, W.W.S.; Hradsky, A.; Kumaraswami, L.; Liakopoulos, V.; Rakhimova, Z.; Saadi, G.; et al. Kidney health for everyone everywhere: From prevention to detection and equitable access to care. Am. J. Hypertens. 2020, 33, 282–289. [Google Scholar] [CrossRef]
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic kidney disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef]
- Foreman, K.J.; Marquez, N.; Dolgert, A.; Fukutaki, K.; Fullman, N.; McGaughey, M.; Pletcher, M.A.; Smith, A.; Tang, K.; Yuan, C.; et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet 2018, 392, 2052–2090. [Google Scholar] [CrossRef]
- Coresh, J.; Selvin, E.; Stevens, L.A.; Manzi, J.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Levey, A.S. Prevalence of chronic kidney disease in the United States. JAMA 2007, 298, 2038–2047. [Google Scholar] [CrossRef]
- Saran, R.; Li, Y.; Robinson, B.; Abbott, K.C.; Agodoa, L.Y.; Ayanian, J.; Bragg-Gresham, J.; Balkrishnan, R.; Chen, J.L.; Cope, E.; et al. US renal data system 2015 annual data report: Epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 2016, 67, A7–A8. [Google Scholar] [CrossRef]
- Levin, A.; Stevens, P.E.; Bilous, R.W.; Coresh, J.; De Francisco, A.L.M.; De Jong, P.E.; Griffith, K.E.; Hemmelgarn, B.R.; Iseki, K.; Edmund, L.; et al. Kidney disease: Improving global outcomes (KDIGO) CKD work group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl. 2013, 3, 1–150. [Google Scholar]
- Forbes, A.; Gallagher, H. Chronic kidney disease in adults: Assessment and management. Clin. Med. 2020, 20, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Jha, V.; Garcia-Garcia, G.; Iseki, K.; Li, Z.; Naicker, S.; Plattner, B.; Saran, R.; Wang, A.Y.; Yang, C.W. Chronic kidney disease: Global dimension and perspectives. Lancet 2013, 382, 260–272. [Google Scholar] [CrossRef]
- Liyanage, T.; Ninomiya, T.; Jha, V.; Neal, B.; Patrice, H.M.; Okpechi, I.; Zhao, M.H.; Lv, J.; Garg, A.X.; Knight, J.; et al. Worldwide access to treatment for end-stage kidney disease: A systematic review. Lancet 2015, 385, 1975–1982. [Google Scholar] [CrossRef]
- Fishman, J.A. Infection in renal transplant recipients. Semin. Nephrol. 2007, 27, 445–461. [Google Scholar] [CrossRef]
- Dantal, J.; Soulillou, J.P. Immunosuppressive drugs and the risk of cancer after organ transplantation. N. Engl. J. Med. 2005, 352, 1371–1373. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C.; Cooper, M.E.; Zimmet, P. Changing epidemiology of type 2 diabetes mellitus and associated chronic kidney disease. Nat. Rev. Nephrol. 2016, 12, 73–81. [Google Scholar] [CrossRef]
- Foley, R.N.; Collins, A.J. The growing economic burden of diabetic kidney disease. Curr. Diabetes Rep. 2009, 9, 460–465. [Google Scholar] [CrossRef]
- Imperatore, G.; Boyle, J.P.; Thompson, T.J.; Case, D.; Dabelea, D.; Hamman, R.F.; Lawrence, J.M.; Liese, A.D.; Liu, L.L.; Mayer-Davis, E.J.; et al. Projections of type 1 and type 2 diabetes burden in the U.S. population aged <20 years through 2050: Dynamic modeling of incidence, mortality, and population growth. Diabetes Care 2012, 35, 2515–2520. [Google Scholar] [CrossRef]
- Haileamlak, A. Chronic kidney disease is on the rise. Ethiop. J. Health Sci. 2018, 28, 681–682. [Google Scholar] [PubMed]
- Barsoum, R.S. Chronic kidney disease in the developing world. N. Engl. J. Med. 2006, 354, 997–999. [Google Scholar] [CrossRef] [PubMed]
- USRDS. 2012 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States; National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2012.
- Thompson, S.; James, M.; Wiebe, N.; Hemmelgarn, B.; Manns, B.; Klarenbach, S.; Tonelli, M. Cause of death in patients with reduced kidney function. J. Am. Soc. Nephrol. 2015, 26, 2504–2511. [Google Scholar] [CrossRef] [PubMed]
- Ronco, C. The Cardiorenal syndrome: Basis and common ground for a multidisciplinary patient-oriented therapy. Cardiorenal Med. 2011, 1, 3–4. [Google Scholar] [CrossRef]
- Gnanaraj, J.; Radhakrishnan, J. Cardio-renal syndrome. F1000Research 2016, 5, 2123. [Google Scholar] [CrossRef]
- Ruggenenti, P.; Cravedi, P.; Remuzzi, G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat. Rev. Nephrol. 2010, 6, 319–330. [Google Scholar] [CrossRef]
- Hou, F.F.; Zhang, X.; Zhang, G.H.; Xie, D.; Chen, P.Y.; Zhang, W.R.; Jiang, J.P.; Liang, M.; Wang, G.B.; Liu, Z.R.; et al. Efficacy and safety of benazepril for advanced chronic renal insufficiency. N. Engl. J. Med. 2006, 354, 131–140. [Google Scholar] [CrossRef]
- Xie, X.; Liu, Y.; Perkovic, V.; Li, X.; Ninomiya, T.; Hou, W.; Zhao, N.; Liu, L.; Lv, J.; Zhang, H.; et al. Renin-angiotensin system inhibitors and kidney and cardiovascular outcomes in patients with CKD: A bayesian network meta-analysis of randomized clinical trials. Am. J. Kidney Dis. 2016, 67, 728–741. [Google Scholar] [CrossRef]
- Brenner, B.M.; Cooper, M.E.; De Zeeuw, D.; Keane, W.F.; Mitch, W.E.; Parving, H.-H.; Remuzzi, G.; Snapinn, S.M.; Zhang, Z.; Shahinfar, S. Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N. Engl. J. Med. 2001, 345, 861–869. [Google Scholar] [CrossRef]
- Zatz, R.; Dunn, B.R.; Meyer, T.W.; Anderson, S.; Rennke, H.G.; Brenner, B.M. Prevention of diabetic glomerulopathy by pharmacological amelioration of glomerular capillary hypertension. J. Clin. Investig. 1986, 77, 1925–1930. [Google Scholar] [CrossRef]
- Holtkamp, F.A.; de Zeeuw, D.; Thomas, M.C.; Cooper, M.E.; de Graeff, P.A.; Hillege, H.J.; Parving, H.-H.; Brenner, B.M.; Shahinfar, S.; Heerspink, H.J.L. An acute fall in estimated glomerular filtration rate during treatment with losartan predicts a slower decrease in long-term renal function. Kidney Int. 2011, 80, 282–287. [Google Scholar] [CrossRef]
- Breyer, M.D.; Susztak, K. Developing treatments for chronic kidney disease in the 21st century. Semin. Nephrol. 2016, 36, 436–447. [Google Scholar] [CrossRef]
- Parving, H.-H.; Brenner, B.M.; Mcmurray, J.; De Zeeuw, D.; Haffner, S.M.; Solomon, S.D.; Chaturvedi, N.; Persson, F.; Desai, A.S.; Nicolaides, M.; et al. Cardiorenal end points in a trial of aliskiren for type 2 diabetes. N. Engl. J. Med. 2012, 367, 2204–2213. [Google Scholar] [CrossRef] [PubMed]
- Fried, L.F.; Emanuele, N.; Zhang, J.H.; Brophy, M.; Conner, T.A.; Duckworth, W.; Leehey, D.J.; McCullough, P.A.; O’Connor, T.; Palevsky, P.; et al. Combined angiotensin inhibition for the treatment of diabetic nephropathy. N. Engl. J. Med. 2013, 369, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Raguraman, P.R.; Kosbar, T.R.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Antisense oligonucleotides targeting angiogenic factors as potential cancer therapeutics. Mol. Ther. Nucleic Acids 2019, 14, 142–157. [Google Scholar] [CrossRef]
- Scoles, D.R.; Minikel, E.V.; Pulst, S.M. Antisense oligonucleotides: A primer. Neurol. Genet. 2019, 5, e323. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77. [Google Scholar] [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Good, L. Translation repression by antisense sequence. Cell Mol. Life Sci. 2003, 60, 854–861. [Google Scholar] [CrossRef]
- Bennett, C.F.; Cowsert, L.M. Antisense oligonucleotides as a tool for gene functionalization and target validation. Biochim. Biophys. Acta 1999, 1489, 19–30. [Google Scholar] [CrossRef]
- Yin, W.; Rogge, M. Targeting RNA: A transformative therapeutic strategy. Clin. Transl. Sci. 2019, 12, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; van Ommen, G.J. Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA 2007, 13, 1609–1624. [Google Scholar] [CrossRef] [PubMed]
- Kurreck, J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003, 270, 1628–1644. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Nakatani, M.; Narukawa, K.; Obika, S. Antisense drug discovery and development. Future Med. Chem. 2011, 3, 339–365. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Sharmab, R.K.; Singh, S.K. Antisense oligonucleotides: Modifications and clinical trials. Med. Chem. Commun. 2014, 5, 1454–1471. [Google Scholar] [CrossRef]
- Wan, W.B.; Seth, P.P. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 2016, 59, 9645–9667. [Google Scholar] [CrossRef]
- Khvorova, A.; Watts, J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017, 35, 238–248. [Google Scholar] [CrossRef]
- Shen, X.; Corey, D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. [Google Scholar] [CrossRef]
- Ghosh, M.K.; Ghosh, K.; Dahl, O.; Cohen, J.S. Evaluation of some properties of a phosphorodithioate oligodeoxyribonucleotide for antisense application. Nucleic Acids Res. 1993, 21, 5761–5766. [Google Scholar] [CrossRef][Green Version]
- Miller, P.S. Oligonucleoside methylphosphonates as antisense reagents. Biotechnology 1991, 9, 358–362. [Google Scholar] [CrossRef]
- Rait, V.; Sergueev, D.; Summers, J.; He, K.; Huang, F.; Krzyzanowska, B.; Shaw, B.R. Boranophosphate nucleic acids—A versatile DNA backbone. Nucleosides Nucleotides 1999, 18, 1379–1380. [Google Scholar] [CrossRef]
- Sheehan, D.; Lunstad, B.; Yamada, C.M.; Stell, B.G.; Caruthers, M.H.; Dellinger, D.J. Biochemical properties of phosphonoacetate and thiophosphonoacetate oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 4109–4118. [Google Scholar] [CrossRef] [PubMed]
- Miroshnichenko, S.K.; Patutina, O.A.; Burakova, E.A.; Chelobanov, B.P.; Fokina, A.A.; Vlassov, V.V.; Altman, S.; Zenkova, M.A.; Stetsenko, D.A. Mesyl phosphoramidate antisense oligonucleotides as an alternative to phosphorothioates with improved biochemical and biological properties. Proc. Natl. Acad. Sci. USA 2019, 116, 1229–1234. [Google Scholar] [CrossRef]
- Patutina, O.A.; Gaponova Miroshnichenko, S.K.; Sen’kova, A.V.; Savin, I.A.; Gladkikh, D.V.; Burakova, E.A.; Fokina, A.A.; Maslov, M.A.; Shmendel, E.V.; Wood, M.; et al. Mesyl phosphoramidate backbone modified antisense oligonucleotides targeting miR-21 with enhanced in vivo therapeutic potency. Proc. Natl. Acad. Sci. USA 2020, 117, 32370–32379. [Google Scholar] [CrossRef]
- Anderson, B.A.; Freestone, G.C.; Low, A.; De-Hoyos, C.L.; Drury, W.J., III; Østergaard, M.E.; Migawa, M.T.; Fazio, M.; Wan, W.B.; Berdeja, A.; et al. Towards next generation antisense oligonucleotides: Mesylphosphoramidate modification improves therapeutic index and duration of effect of gapmer antisense oligonucleotides. Nucleic Acids Res. 2021, 49, 9026–9041. [Google Scholar] [CrossRef] [PubMed]
- Majlessi, M.; Nelson, N.C.; Becker, M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998, 26, 2224–2229. [Google Scholar] [CrossRef]
- Miroshnichenko, S.K.; Amirloo, B.; Bichenkova, E.V.; Vlassov, V.V.; Zenkova, M.A.; Patutina, O.A. 2’OMe modification of antimirna-21 oligonucleotide–peptide conjugate improves its hybridization properties and catalytic activity. Russ. J. Bioorg. Chem. 2019, 45, 803–812. [Google Scholar] [CrossRef]
- Geary, R.S.; Watanabe, T.A.; Truong, L.; Freier, S.; Lesnik, E.A.; Sioufi, N.B.; Sasmor, H.; Manoharan, M.; Levin, A.A. Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 2001, 296, 890–897. [Google Scholar] [PubMed]
- Koizumi, M.; Takagi-Sato, M.; Okuyama, R.; Araki, K.; Sun, W.; Nakai, D.; Tsutsumi, S.; Kawai, K. Direct comparison of in vivo antisense activity of ENA oligonucleotides targeting ptp1b mRNA with that of 2′-O-(2-methoxy) ethyl-modified oligonucleotides. Oligonucleotides 2006, 16, 253–262. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked nucleic acids: Promising nucleic acid analogs for therapeutic applications. Chem. Biodivers. 2010, 7, 536–542. [Google Scholar] [CrossRef]
- Le, B.T.; Adams, A.M.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Rational design of short locked nucleic acid-modified 2′-O-methyl antisense oligonucleotides for efficient exon-skipping in vitro. Mol. Ther. Nucleic Acids 2017, 9, 155–161. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009, 6, 321–323. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, S.; Chaput, J.C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012, 4, 183–187. [Google Scholar] [CrossRef]
- Lin, Y.; Qiu, Q.; Gill, S.C.; Jayasena, S.D. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 1994, 22, 5229–5234. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Le, B.T.; Chakravarthy, M.; Kosbar, T.R.; Veedu, R.N. Systematic evaluation of 2′-fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Alves Ferreira-Bravo, I.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Chen, S.; Abramov, M.; Herdewijn, P.; Veedu, R.N. Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and d-altritol nucleic acid-modified 2′-O-methyl RNA mixmer antisense oligonucleotides for exon skipping in vitro. Chem. Commun. 2016, 52, 13467–13470. [Google Scholar] [CrossRef]
- Pallan, P.S.; Allerson, C.R.; Berdeja, A.; Seth, P.P.; Swayze, E.E.; Prakash, T.P.; Egli, M. Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs. Chem. Comm. 2012, 48, 8195–8197. [Google Scholar] [CrossRef] [PubMed]
- Hyrup, B.; Nielsen, P.E. Peptide nucleic acids (PNA): Synthesis, properties and potential applications. Bioorg. Med. Chem. 1996, 4, 5–23. [Google Scholar] [CrossRef]
- Summerton, J.; Weller, D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997, 7, 187–195. [Google Scholar] [CrossRef]
- Le, B.T.; Veedu, R.N.; Fletcher, S.; Wilton, S.D. Antisense oligonucleotide development for the treatment of muscular dystrophies. Expert Opin. Orphan Drugs 2016, 4, 139–152. [Google Scholar]
- Agrawal, S.; Jiang, Z.; Zhao, Q.; Shaw, D.; Cai, Q.; Roskey, A.; Channavajjala, L.; Saxinger, C.; Zhang, R. Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: In vitro and in vivo studies. Proc. Natl. Acad. Sci. USA 1997, 94, 2620–2625. [Google Scholar] [CrossRef]
- Stanton, R.; Sciabola, S.; Salatto, C.; Weng, Y.; Moshinsky, D.; Little, J.; Walters, E.; Kreeger, J.; DiMattia, D.; Chen, T.; et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012, 22, 344–359. [Google Scholar] [CrossRef]
- Monia, B.P.; Lesnik, E.A.; Gonzalez, C.; Lima, W.F.; McGee, D.; Guinosso, C.J.; Kawasaki, A.M.; Cook, P.D.; Freier, S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar] [CrossRef]
- Chen, S.; Le, B.T.; Rahimizadeh, K.; Shaikh, K.; Mohal, N.; Veedu, R.N. Synthesis of a morpholino nucleic acid (MNA)-uridine phosphoramidite, and exon skipping using MNA/2′-O-methyl mixmer antisense oligonucleotide. Molecules 2016, 21, 1582. [Google Scholar] [CrossRef] [PubMed]
- Langner, H.K.; Jastrzebska, K.; Caruthers, M.H. Synthesis and characterization of thiophosphoramidate morpholino oligonucleotides and chimeras. J. Am. Chem. Soc. 2020, 142, 16240–16253. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.K.; Corey, D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012, 226, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Siomi, M.C. On the road to reading the RNA-interference code. Nature 2009, 457, 396–404. [Google Scholar] [CrossRef]
- Sajid, M.I.; Moazzam, M.; Kato, S.; Yeseom Cho, K.; Tiwari, R.K. Overcoming barriers for siRNA therapeutics: From bench to bedside. Pharmaceuticals 2020, 13, 294. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal. Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Jackson, A.L.; Linsley, P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010, 9, 57–67. [Google Scholar] [CrossRef]
- Jackson, A.L.; Burchard, J.; Leake, D.; Reynolds, A.; Schelter, J.; Guo, J.; Johnson, J.M.; Lim, L.; Karpilow, J.; Nichols, K.; et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 2006, 12, 1197–1205. [Google Scholar] [CrossRef]
- Balachandran, A.A.; Larcher, L.M.; Chen, S.; Veedu, R.N. Therapeutically significant microRNAs in primary and metastatic brain malignancies. Cancers 2020, 12, 2534. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Piva, R.; Spandidos, D.A.; Gambari, R. From microRNA functions to microRNA therapeutics: Novel targets and novel drugs in breast cancer research and treatment (review). Int. J. Oncol. 2013, 43, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Wang, S.C.; Hsu, C.Y.; Miao, Y.; Martin, M.; Yin, Y.; Wu, C.C.; Wang, Y.T.; Wu, G.; Chien, S.; et al. MicroRNA-92a mediates endothelial dysfunction in CKD. J. Am. Soc. Nephrol. 2017, 28, 3251–3261. [Google Scholar] [CrossRef] [PubMed]
- Hinkel, R.; Penzkofer, D.; Zühlke, S.; Fischer, A.; Husada, W.; Xu, Q.F.; Baloch, E.; van Rooij, E.; Zeiher, A.M.; Kupatt, C.; et al. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation 2013, 128, 1066–1075. [Google Scholar] [CrossRef]
- Moreno, J.A.; Hamza, E.; Guerrero-Hue, M.; Rayego-Mateos, S.; García-Caballero, C.; Vallejo-Mudarra, M.; Metzinger, L.; Meuth, V.M.-L. Non-coding RNAs in kidney diseases: The long and short of them. Int. J. Mol. Sci. 2021, 22, 6077. [Google Scholar] [CrossRef]
- Li, N.; Cui, Y.; Yin, M.; Liu, F. Screening potential prognostic biomarkers of long non-coding RNAs for predicting the risk of chronic kidney disease. Braz. J. Med. Biol. Res. 2019, 52, e8333. [Google Scholar] [CrossRef]
- Daniel, C.; Takabatake, Y.; Mizui, M.; Isaka, Y.; Kawashi, H.; Rupprecht, H.; Imai, E.; Hugo, C. Antisense oligonucleotides against thrombospondin-1 inhibit activation of TGF- β in fibrotic renal disease in the rat in vivo. Am. J. Pathol. 2003, 163, 1185–1192. [Google Scholar] [CrossRef]
- Guha, M.; Xu, Z.G.; Tung, D.; Lanting, L.; Natarajan, R. Specific down-regulation of connective tissue growth factor attenuates progression of nephropathy in mouse models of type 1 and type 2 diabetes. FASEB J. 2007, 21, 3355–3368. [Google Scholar] [CrossRef]
- Wang, J.H.; Newbury, L.J.; Knisely, A.S.; Monia, B.; Hendry, B.M.; Sharpe, C.C. Antisense knockdown of Kras inhibits fibrosis in a rat model of unilateral ureteric obstruction. Am. J. Pathol. 2012, 180, 82–90. [Google Scholar] [CrossRef]
- Ravichandran, K.; Zafar, I.; He, Z.; Doctor, R.B.; Moldovan, R.; Mullick, A.E.; Edelstein, C.L. An mTOR anti-sense oligonucleotide decreases polycystic kidney disease in mice with a targeted mutation in Pkd2. Hum. Mol. Genet. 2014, 23, 4919–4931. [Google Scholar] [CrossRef]
- Ravichandran, K.; Ozkok, A.; Wang, Q.; Mullick, A.E.; Edelstein, C.L. Antisense-mediated angiotensinogen inhibition slows polycystic kidney disease in mice with a targeted mutation in Pkd2. Am. J. Physiol. Ren. Physiol. 2015, 308, F349–F357. [Google Scholar] [CrossRef]
- Aghajan, M.; Booten, S.L.; Althage, M.; Hart, C.E.; Ericsson, A.; Maxvall, I.; Ochaba, J.; Menschik-Lundin, A.; Hartleib, J.; Kuntz, S.; et al. Antisense oligonucleotide treatment ameliorates IFN-γ-induced proteinuria in APOL1-transgenic mice. JCI Insight 2019, 4, e126124. [Google Scholar] [CrossRef] [PubMed]
- Bülow, R.D.; Boor, P. Extracellular matrix in kidney fibrosis: More than just a scaffold. J. Histochem. Cytochem. 2019, 67, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Border, W.A.; Noble, N.A. Transforming growth factor-β in tissue fibrosis. N. Engl. J. Med. 1994, 331, 1286–1292. [Google Scholar] [PubMed]
- Okuda, S.; Languino, L.R.; Rouslahti, E.; Border, W.A. Elevated expression of transforming growth factor-β and proteoglycan production in experimental glomerulonephritis. J. Clin. Investig. 1990, 86, 453–462. [Google Scholar] [CrossRef]
- Kopp, J.B.; Factor, V.M.; Mozes, M.; Nagy, P.; Sanderson, N.; Böttinger, E.P.; Klotman, P.E.; Thorgeirsson, S.S. Transgenic mice with increased plasma levels of TGF-β1 develop progressive renal disease. Lab. Investig. 1996, 74, 991–1003. [Google Scholar]
- Sharma, K.; Ziyadeh, F.N. The emerging role of transforming growth factor-β in kidney disease. Am. J. Physiol. 1994, 35, F829–F842. [Google Scholar] [CrossRef]
- Akagi, Y.; Isaka, Y.; Arai, M.; Kaneko, T.; Takenaka, M.; Moriyama, T.; Kaneda, Y.; Ando, A.; Orita, Y.; Kamada, T.; et al. Inhibition of TGF-β 1 expression by antisense oligonucleotides suppressed extracellular matrix accumulation in experimental glomerulonephritis. Kidney Int. 1996, 50, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Shull, M.M.; Ormsby, I.; Kier, A.B.; Pawlowski, S.; Diebold, R.J.; Yin, M.; Allen, R.; Sidman, C.; Proetzel, G.; Calvin, D.; et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 1992, 359, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Sanford, L.P.; Ormsby, I.; Gittenberger-de Groot, A.; Sariola, H.; Friedman, R.; Boivin, G.P.; Cardell, E.L.; Doetschmann, T. TGF-β2 knockout mice have multiple developmental defects that are nonoverlapping with other TGF-β knockout phenotypes. Development 1997, 124, 2659–2670. [Google Scholar] [CrossRef] [PubMed]
- Kaartinen, V.; Voncken, J.W.; Shuler, C.; Warbuton, D.; Bu, D.; Heitserkamp, N.; Groffen, J. Abnormal lung development and cleft palate in mice lacking TGF-β3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995, 11, 415–421. [Google Scholar] [CrossRef]
- Schultz-Cherry, S.; Murphy-Ullrich, J.E. Thrombospondin causes activation of latent transforming growth factor-β secreted by endothelial cells by a novel mechanism. J. Cell Biol. 1993, 122, 923–932. [Google Scholar] [CrossRef]
- Tada, H.; Isogai, S. The fibronectin production is increased by thrombospondin via activation of TGF-β in cultured human mesangial cells. Nephron 1998, 79, 38–44. [Google Scholar] [CrossRef]
- Crawford, S.E.; Chen, H.; Mosher, D.; Misenheimer, T.; Krutzsch, H.; Roberts, D.D.; Murphy-Ullrich, J.E. Thrombospondin-1 is a major activator of TGF-β1 in vivo. Cell 1998, 93, 1159–1170. [Google Scholar] [CrossRef]
- Lawler, J.; Sunday, M.; Thibert, V.; Duquette, M.; George, E.L.; Rayburn, H.; Hynes, R.O. Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J. Clin. Investig. 1998, 101, 982–992. [Google Scholar] [CrossRef]
- Hugo, C.; Shankland, S.J.; Pichler, R.H.; Couser, W.G.; Johnson, R.J. Thrombospondin 1 precedes and predicts the development of tubulointerstitial fibrosis in glomerular disease in the rat. Kidney Int. 1998, 53, 302–311. [Google Scholar] [CrossRef]
- Hugo, C.; Pichler, R.; Meek, R.; Gordon, K.; Kyriakides, T.; Floege, J.; Bornstein, P.; Couser, W.; Johnson, R.J. Thrombospondin1 is expressed by proliferating mesangial cells in vivo and is up-regulated by PDGF and bFGF. Kidney Int. 1995, 48, 1846–1856. [Google Scholar] [CrossRef]
- Kreisberg, J.I.; Ayo, S.H. The glomerular mesangium in diabetes mellitus. Kidney Int. 1993, 43, 109–113. [Google Scholar] [CrossRef][Green Version]
- Ziyadeh, F.N.; Sharma, K.; Ericksen, M.; Wolf, G. Stimulation of collagen gene expression and protein synthesis in murine mesangial cells by high glucose is mediated by autocrine activation of transforming growth factor-β. J. Clin. Investig. 1994, 93, 536–542. [Google Scholar] [CrossRef]
- Ziyadeh, F.N.; Han, D.C. Involvement of transforming growth factor-beta and its receptors in the pathogenesis of diabetic nephrology. Kidney Int. 1997, 60, S7–S11. [Google Scholar]
- Hoffman, B.B.; Sharma, K.; Zhu, Y.; Ziyadeh, F.N. Transcriptional activation of transforming growth factor-β1 in mesangial cell culture by high glucose concentration. Kidney Int. 1998, 54, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Han, D.C.; Isono, M.; Hoffman, B.B.; Ziyadeh, F.N. High glucose stimulates proliferation and collagen type I synthesis in renal cortical fibroblasts: Mediation by autocrine activation of TGF-beta. J. Am. Soc. Nephrol. 1999, 10, 1891–1899. [Google Scholar] [CrossRef]
- Reeves, W.B.; Andreoli, T.E. Transforming growth factor beta contributes to progressive diabetic nephropathy. Proc. Natl. Acad. Sci. USA 2000, 97, 7667–7669. [Google Scholar] [CrossRef] [PubMed]
- Riser, B.L.; Denichilo, M.; Cortes, P.; Baker, C.; Grondin, J.M.; Yee, J.; Narins, R.G. Regulation of connective tissue growth factor activity in cultured rat mesangial cells and its expression in experimental diabetic glomerulosclerosis. J. Am. Soc. Nephrol. 2000, 11, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Sakharova, O.V.; Taal, M.W.; Brenner, B.M. Pathogenesis of diabetic nephropathy: Focus on transforming growth factor-beta and connective tissue growth factor. Curr. Opin. Nephrol. Hypertens. 2001, 10, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.; Godson, C.; Cannon, S.; Kato, S.; Mackenzie, H.S.; Martin, F.; Brady, H.R. Suppression subtractive hybridization identifies high glucose levels as a stimulus for expression of connective tissue growth factor and other genes in human mesangial cells. J. Biol. Chem. 1999, 274, 5830–5834. [Google Scholar] [CrossRef]
- Okada, H.; Kikuta, T.; Kobayashi, T.; Inoue, T.; Kanno, Y.; Takigawa, T.; Sugaya, T.; Kopp, J.B.; Suzuki, H. Connective tissue growth factor expressed in tubular epithelium plays a pivotal role in renal fibrogenesis. J. Am. Soc. Nephrol. 2005, 16, 133–143. [Google Scholar] [CrossRef]
- Norman, J.T.; Fine, L.G. Progressive renal disease: Fibroblasts, extracellular matrix, and integrins. Exp. Nephrol. 1999, 7, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Wittinghofer, A.; Scheffzek, K.; Ahmadian, M.R. The interaction of Ras with GTPase-activating proteins. FEBS Lett. 1997, 410, 63–67. [Google Scholar] [CrossRef]
- Bechtel, W.; McGoohan, S.; Zeisberg, E.M.; Muller, G.A.; Kalbacher, H.; Salant, D.J.; Muller, C.A.; Kalluri, R.; Zeisberg, M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 2010, 16, 544–550. [Google Scholar] [CrossRef]
- Janda, E.; Lehmann, K.; Killisch, I.; Jechlinger, M.; Herzig, M.; Downward, J.; Beug, H.; Grunert, S. Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis: Dissection of Ras signaling pathways. J. Cell Biol. 2002, 156, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.C.; Kocher, H.M.; Khwaja, A.; Kloog, Y.; Cook, H.T.; Hendry, B.M. Ras antagonist farnesylthiosalicylic acid (FTS) reduces glomerular cellular proliferation and macrophage number in rat thy-1 nephritis. J. Am. Soc. Nephrol. 2003, 14, 848–854. [Google Scholar] [CrossRef]
- Kocher, H.M.; Moorhead, J.; Sharpe, C.C.; Dockrell, M.E.; Al-Nawab, M.; Hendry, B.M. Expression of Ras GTPases in normal kidney and in glomerulonephritis. Nephrol. Dial. Transplant. 2003, 18, 2284–2292. [Google Scholar] [CrossRef]
- Martinez-Salgado, C.; Fuentes-Calvo, I.; Garcia-Cenador, B.; Santos, E.; Lopez-Novoa, J.M. Involvement of H- and N-Ras isoforms in transforming growth factor-β1-induced proliferation and in collagen and fibronectin synthesis. Exp. Cell Res. 2006, 312, 2093–2106. [Google Scholar] [CrossRef]
- Rodriguez-Pena, A.B.; Grande, M.T.; Eleno, N.; Arevalo, M.; Guerrero, C.; Santos, E.; Lopez-Novoa, J.M. Activation of Erk1/2 and Akt following unilateral ureteral obstruction. Kidney Int. 2008, 74, 196–209. [Google Scholar] [CrossRef]
- Dockrell, M.E.; Phanish, M.K.; Hendry, B.M. Tgf-β auto-induction and connective tissue growth factor expression in human renal tubule epithelial cells requires N-ras. Nephron Exp. Nephrol. 2009, 112, e71–e79. [Google Scholar] [CrossRef]
- Lahsnig, C.; Mikula, M.; Petz, M.; Zulehner, G.; Schneller, D.; van Zijl, F.; Huber, H.; Csiszar, A.; Beug, H.; Mikulits, W. ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene 2009, 28, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Grande, M.T.; Fuentes-Calvo, I.; Arevalo, M.; Heredia, F.; Santos, E.; Martinez-Salgado, C.; Rodriguez-Puyol, D.; Nieto, M.A.; Lopez-Novoa, J.M. Deletion of H-Ras decreases renal fibrosis and myofibroblast activation following ureteral obstruction in mice. Kidney Int. 2010, 77, 509–518. [Google Scholar] [CrossRef]
- Sharpe, C.C.; Dockrell, M.E.; Noor, M.I.; Monia, B.P.; Hendry, B.M. Role of Ras isoforms in the stimulated proliferation of human renal fibroblasts in primary culture. J. Am. Soc. Nephrol. 2000, 11, 1600–1606. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, C.C.; Dockrell, M.E.; Scott, R.; Noor, M.I.; Cowsert, L.M.; Monia, B.P.; Hendry, B.M. Evidence of a role for Ki-Ras in the stimulated proliferation of renal fibroblasts. J. Am. Soc. Nephrol. 1999, 10, 1186–1192. [Google Scholar] [CrossRef]
- Klahr, S.; Morrissey, J. Obstructive nephropathy and renal fibrosis. Am. J. Physiol. Ren. Physiol. 2002, 283, F861–F875. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.J.; Revenko, A.S.; Hanson, L.L.; Ellston, R.; Staniszewska, A.; Whalley, N.; Pandey, S.K.; Revill, M.; Rooney, C.; Buckett, L.K.; et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017, 9, eaal5253. [Google Scholar] [CrossRef]
- Grantham, J.J. Clinical practice. Autosomal dominant polycystic kidney disease. N. Engl. J. Med. 2008, 359, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, C.L. Mammalian target of rapamycin and caspase inhibitors in polycystic kidney disease. Clin. J. Am. Soc. Nephrol. 2008, 3, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Shillingford, J.M.; Murcia, N.S.; Larson, C.H.; Low, S.H.; Hedgepeth, R.; Brown, N.; Flask, C.A.; Novick, A.C.; Goldfarb, D.A.; Kramer-Zucker, A.; et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2006, 103, 5466–5471. [Google Scholar] [CrossRef]
- Shillingford, J.M.; Piontek, K.B.; Germino, G.G.; Weimbs, T. Rapamycin ameliorates PKD resulting from conditional inactivation of Pkd1. J. Am. Soc. Nephrol. 2010, 21, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Wahl, P.R.; Serra, A.L.; Le Hir, M.; Molle, K.D.; Hall, M.N.; Wuthrich, R.P. Inhibition of mTOR with sirolimus slows disease progression in Han:SPRD rats with autosomal dominant polycystic kidney disease (ADPKD). Nephrol. Dial. Transplant. 2006, 21, 598–604. [Google Scholar] [CrossRef]
- Fischer, D.C.; Jacoby, U.; Pape, L.; Ward, C.J.; Kuwertz-Broeking, E.; Renken, C.; Nizze, H.; Querfeld, U.; Rudolph, B.; Mueller-Wiefel, D.E.; et al. Activation of the AKT/mTOR pathway in autosomal recessive polycystic kidney disease (ARPKD). Nephrol. Dial. Transplant. 2009, 24, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef]
- Rodrik-Outmezguine, V.S.; Chandarlapaty, S.; Pagano, N.C.; Poulikakos, P.I.; Scaltriti, M.; Moskatel, E.; Baselga, J.; Guichard, S.; Rosen, N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011, 1, 248–259. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Sengupta, S.; Sheen, J.H.; Hsu, P.P.; Bagley, A.F.; Markhard, A.L.; Sabatini, D.M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 2006, 22, 159–168. [Google Scholar] [CrossRef]
- Belibi, F.; Ravichandran, K.; Zafar, I.; He, Z.; Edelstein, C.L. mTORC1/2 and rapamycin in female Han:SPRD rats with polycystic kidney disease. Am. J. Physiol. 2011, 300, F236–F244. [Google Scholar] [CrossRef]
- Natoli, T.A.; Smith, L.A.; Rogers, K.A.; Wang, B.; Komarnitsky, S.; Budman, Y.; Belenky, A.; Bukanov, N.O.; Dackowski, W.R.; Husson, H.; et al. Inhibition of glucosylceramide accumulation results in effective blockade of polycystic kidney disease in mouse models. Nat. Med. 2010, 16, 788–792. [Google Scholar] [CrossRef]
- Zafar, I.; Ravichandran, K.; Belibi, F.; Doctor, R.B.; Edelstein, C.L. Sirolimus attenuates disease progression in an orthologous mouse model of human autosomal dominant polycystic kidney disease. Kidney Int. 2010, 78, 754–761. [Google Scholar] [CrossRef]
- Serra, A.L.; Poster, D.; Kistler, A.D.; Krauer, F.; Raina, S.; Young, J.; Rentsch, K.M.; Spanaus, K.S.; Senn, O.; Kristanto, P.; et al. Sirolimus and kidney growth in autosomal dominant polycystic kidney disease. N. Engl. J. Med. 2010, 363, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Shor, B.; Gibbons, J.J.; Abraham, R.T.; Yu, K. Targeting mTOR globally in cancer; thinking beyond rapamycin. Cell Cycle 2009, 8, 3831–3837. [Google Scholar] [CrossRef] [PubMed]
- Dazert, E.; Hall, M.N. mTOR signaling in disease. Curr. Opin. Cell Biol. 2011, 23, 744–755. [Google Scholar] [CrossRef]
- Wilson, P.D. Polycystic kidney disease. N. Engl. J. Med. 2004, 350, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Raizada, V.; Skipper, B.; Luo, W.; Griffith, J. Intracardiac and intrarenal renin-angiotensin systems: Mechanisms of cardiovascular and renal effects. J. Investig. Med. 2007, 55, 341–359. [Google Scholar] [CrossRef]
- Ruiz-Ortega, M.; Ruperez, M.; Esteban, V.; Rodriguez-Vita, J.; Sanchez-Lopez, E.; Carvajal, G.; Egido, J. Angiotensin II: A key factor in the inflammatory and fibrotic response in kidney diseases. Nephrol. Dial. Transplant. 2006, 21, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Schrier, R.W. Renal volume, renin-angiotensin-aldosterone system, hypertension, and left ventricular hypertrophy in patients with autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2009, 20, 1888–1893. [Google Scholar] [CrossRef]
- Wu, C.; Lu, H.; Cassis, L.A.; Daugherty, A. Molecular and pathophysiological features of angiotensinogen: A mini review. Am. J. Med. Sci. 2011, 4, 183–190. [Google Scholar] [CrossRef]
- Ronco, C.; Di Lullo, L. Cardiorenal syndrome. Heart Fail. Clin. 2014, 10, 251–280. [Google Scholar] [CrossRef]
- Long, D.A.; Price, K.L.; Herrera-Acosta, J.; Johnson, R.J. How does angiotensin II cause renal injury? Hypertension 2004, 43, 722–723. [Google Scholar] [CrossRef]
- Sparks, M.A.; Crowley, S.D.; Gurley, S.B.; Mirotsou, M.; Coffman, T.M. Classical renin-angiotensin system in kidney physiology. Compr. Physiol. 2014, 4, 1201–1228. [Google Scholar]
- Theuer, J.; Dechend, R.; Muller, D.N.; Park, J.K.; Fiebeler, A.; Barta, P.; Ganten, D.; Haller, H.; Dietz, R.; Luft, F.C. Angiotensin II induced inflammation in the kidney and in the heart of double transgenic rats. BMC Cardiovasc. Disord. 2002, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.E.; Malik, H.S. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009, 19, 850–858. [Google Scholar] [CrossRef]
- Monajemi, H.; Fontijn, R.D.; Pannekoek, H.; Horrevoets, A.J. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 2002, 79, 539–546. [Google Scholar] [CrossRef]
- Shukha, K.; Mueller, J.L.; Chung, R.T.; Curry, M.P.; Friedman, D.J.; Pollak, M.R.; Berg, A.H. Most ApoL1 is secreted by the liver. J. Am. Soc. Nephrol. 2017, 28, 1079–1083. [Google Scholar] [CrossRef]
- Vanhamme, L.; Paturiaux-Hanocq, F.; Poelvoorde, P.; Nolan, D.P.; Lins, L.; Van Den Abbeele, J.; Pays, A.; Tebabi, P.; Van Xong, H.; Jacquet, A.; et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 2003, 422, 83–87. [Google Scholar] [CrossRef]
- Pérez-Morga, D.; Vanhollebeke, B.; Paturiaux-Hanocq, F.; Nolan, D.P.; Lins, L.; Homblé, F.; Vanhamme, L.; Tebabi, P.; Pays, A.; Poelvoorde, P.; et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science 2005, 309, 469–472. [Google Scholar] [CrossRef]
- Genovese, G.; Friedman, D.J.; Ross, M.D.; Lecordier, L.; Uzureau, P.; Freedman, B.I.; Bowden, D.W.; Langefeld, C.D.; Oleksyk, T.K.; Uscinski Knob, A.L.; et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010, 329, 841–845. [Google Scholar] [CrossRef]
- Nichols, B.; Jog, P.; Lee, J.H.; Blackler, D.; Wilmot, M.; D’Agati, V.; Markowitz, G.; Kopp, J.B.; Alper, S.L.; Pollak, M.R.; et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015, 87, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Beckerman, P.; Bi-Karchin, J.; Park, A.S.; Qiu, C.; Dummer, P.D.; Soomro, I.; Boustany-Kari, C.M.; Pullen, S.S.; Miner, J.H.; Hu, C.A.; et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat. Med. 2017, 23, 429–438. [Google Scholar] [CrossRef]
- Tzur, S.; Rosset, S.; Shemer, R.; Yudkovsky, G.; Selig, S.; Tarekegn, A.; Bekele, E.; Bradman, N.; Wasser, W.G.; Behar, D.M.; et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet. 2010, 128, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Kopp, J.B.; Nelson, G.W.; Sampath, K.; Johnson, R.C.; Genovese, G.; An, P.; Friedman, D.; Briggs, W.; Dart, R.; Korbet, S.; et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol. 2011, 22, 2129–2137. [Google Scholar] [CrossRef]
- Saran, R.; Robinson, B.; Abbott, K.C.; Agodoa, L.Y.; Albertus, P.; Ayanian, J.; Balkrishnan, R.; Bragg-Gresham, J.; Cao, J.; Chen, J.L.; et al. US renal data system 2016 annual data report: Epidemiology of kidney disease in the United States. Am. J. Kidney Dis. 2017, 69, A7–A8. [Google Scholar] [CrossRef]
- Parsa, A.; Kao, W.H.; Xie, D.; Astor, B.C.; Li, M.; Hsu, C.Y.; Feldman, H.I.; Parekh, R.S.; Kusek, J.W.; Greene, T.H.; et al. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013, 369, 2183–2196. [Google Scholar] [CrossRef]
- Johnstone, D.B.; Shegokar, V.; Nihalani, D.; Rathore, Y.S.; Mallik, L.; Ashish; Zare, V.; Ikizler, H.O.; Powar, R.; Holzman, L.B. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS ONE 2012, 7, e51546. [Google Scholar]
- Joshi, P.P.; Shegokar, V.R.; Powar, R.M.; Herder, S.; Katti, R.; Salkar, H.R.; Dani, V.S.; Bhargava, A.; Jannin, J.; Truc, P. Human trypanosomiasis caused by Trypanosoma evansi in India: The first case report. Am. J. Trop. Med. Hyg. 2005, 73, 491–495. [Google Scholar] [CrossRef]
- Vanhollebeke, B.; Truc, P.; Poelvoorde, P.; Pays, A.; Joshi, P.P.; Katti, R.; Jannin, J.G.; Pays, E. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-I. N. Engl. J. Med. 2006, 355, 2752–2756. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef] [PubMed]
- Asami, Y.; Yoshioka, K.; Nishina, K.; Nagata, T.; Yokota, T. Drug delivery system of therapeutic oligonucleotides. Drug Discov. Ther. 2016, 10, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yang, Z.; Tang, X. Chemical modifications of nucleic acid drugs and their delivery systems for gene-based therapy. Med. Res. Rev. 2018, 38, 829–869. [Google Scholar] [CrossRef]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef]
- Kaczmarek, J.C.; Kowalski, P.S.; Anderson, D.G. Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.I.; Kimura, B. Gene therapy for hypertension: Antisense inhibition of the renin-angiotensin system. Methods Mol. Med. 2005, 108, 363–379. [Google Scholar] [PubMed]
- Fiorentino, M.; Grandaliano, G.; Gesualdo, L.; Castellano, G. Acute kidney injury to chronic kidney disease transition. Contrib. Nephrol. 2018, 193, 45–54. [Google Scholar] [PubMed]
- Ko, G.J.; Grigoryev, D.N.; Linfert, D.; Jang, H.R.; Watkins, T.; Cheadle, C.; Racusen, L.; Rabb, H. Transcriptional analysis of kidneys during repair from AKI reveals possible roles for NGAL and KIM-1 as biomarkers of AKI-to-CKD transition. Am. J. Physiol. Ren. Physiol. 2010, 298, F1472–F1483. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).