1. Introduction
Monitoring the evolution of critical inflammatory conditions is fundamental in the intensive care unit (ICU). In this context, the role of biomarkers is crucial, as they enable clinicians to track disease progression and anticipate clinical deterioration [
1]. In recent years, a new biomarker has emerged in the literature and in clinical practice: the Pancreatic Stone Protein (PSP). Initially, researchers focused on the role of this protein in the management of sepsis, a condition that represents one of the main issues in ICU and is associated with a high rate of mortality due to altered immunological response to infection [
2]. Eggimann et al., in a review, underlined the positive role of PSP in the diagnosis of sepsis, reporting a higher sensibility and specificity than other biomarkers commonly used for this purpose, such as Procalcitonin, C-reactive protein or various cytokines [
3]. In addition, Keel et al. proved that this increases in this biomarker occurred earlier than others in the initial development of this clinical condition [
4]. Klein et al. investigated the role of PSP in patients submitted to cardiac surgery, proving a strong correlation between high protein levels and the risk of developing an infectious complication during hospitalization [
5]. The role of PSP in prognostic evaluation is also crucial. High protein levels are associated with more severe infections and higher mortality rates, as proved by Zuercher et al. [
6]. Finally, recent studies suggest potential diagnostic and prognostic roles for PSP in other infections such as pneumonia, abdominal infections, or even COVID-19 [
7,
8]. Notably, PSP levels increase earlier and more consistently in the presence of bacterial triggers, showing high sensitivity in different clinical settings including postoperative care, trauma, and respiratory infections. However, its role in the context of COVID-19-related sepsis remains under-investigated. In the context of heterogeneous ICU populations and overlapping clinical signs, multivariate prediction tools are increasingly required to support clinical decision-making. To address this, we applied a Least Absolute Shrinkage and Selection Operator (LASSO) regression model, a machine learning technique capable of handling multicollinearity and of selecting the most informative predictors among numerous variables. The aim of this study was to evaluate the performance of PSP and CRP as biomarkers for predicting bacterial infections in critically ill COVID-19 patients, using LASSO and mixed-effects models to ensure statistical robustness. A secondary objective was to explore their association with severe respiratory failure, as expressed by a Horowitz Index (HI) lower than 100. We hypothesized that PSP could serve as an early and reliable indicator of infection, contributing to better clinical risk stratification and antibiotic stewardship in COVID-19 ICU care.
2. Materials and Methods
2.1. Study Design and Setting
This prospective observational study was conducted between March and June 2021 in the COVID-19 Intensive Care Unit (ICU) of the Civil Hospital of Baggiovara (Modena, Italy). The study was approved by the local Ethics Committee (reference number 784/2021). Informed consent was obtained from all patients or their legal representatives, in accordance with Italian regulations.
2.2. Inclusion and Exclusion Criteria
Patients were eligible for inclusion if they were (1) ≥18 years old; (2) admitted to the ICU with confirmed COVID-19; (3) expected to require mechanical ventilation for >24 h; (4) equipped with an arterial catheter; and (5) able to provide informed consent directly or through a legal representative. Exclusion criteria included refusal of consent, presence of a do-not-resuscitate (DNR) order, or previous ICU admission for COVID-19.
2.3. Data Collection
Demographic and clinical data were collected upon ICU admission and included type of respiratory support, vital signs (arterial pressure via invasive monitoring), daily urine output, vasopressor/inotrope use, and prescribed therapies. Blood samples were drawn at admission (T0), and on days 3 (T1), 5 (T2), 7 (T3), 10 (T4), 15 (T5), and 20 (T6). Bacterial infections were diagnosed based on Sepsis-3 criteria [
2], confirmed by positive microbiological cultures (respiratory tract, blood, or urinary tract). Mortality was assessed at 30 days post-admission.
2.4. Biomarker Measurement
PSP levels were measured using nanofluidic immunoassay with the abioSCOPE® system (Abionic SA, Épalinges, Switzerland). CRP and Procalcitonin (PCT) were assessed via standard laboratory techniques. The Horowitz Index (PaO2/FiO2) and Sequential Organ Failure Assessment (SOFA) score were calculated at the same time points.
2.5. Antibiotic Therapy and Stewardship
Antibiotic therapy was administered in accordance with local antimicrobial stewardship protocols based on the national guidelines provided at the time of the study [
9,
10,
11]. Empirical therapy was initiated based on clinical suspicion and microbiological data, and subsequently adjusted following culture results, biomarker trends, and clinical course. Broad-spectrum antibiotics were de-escalated when possible, and therapy duration was tailored according to response and site of infection.
2.6. Statistical Analysis
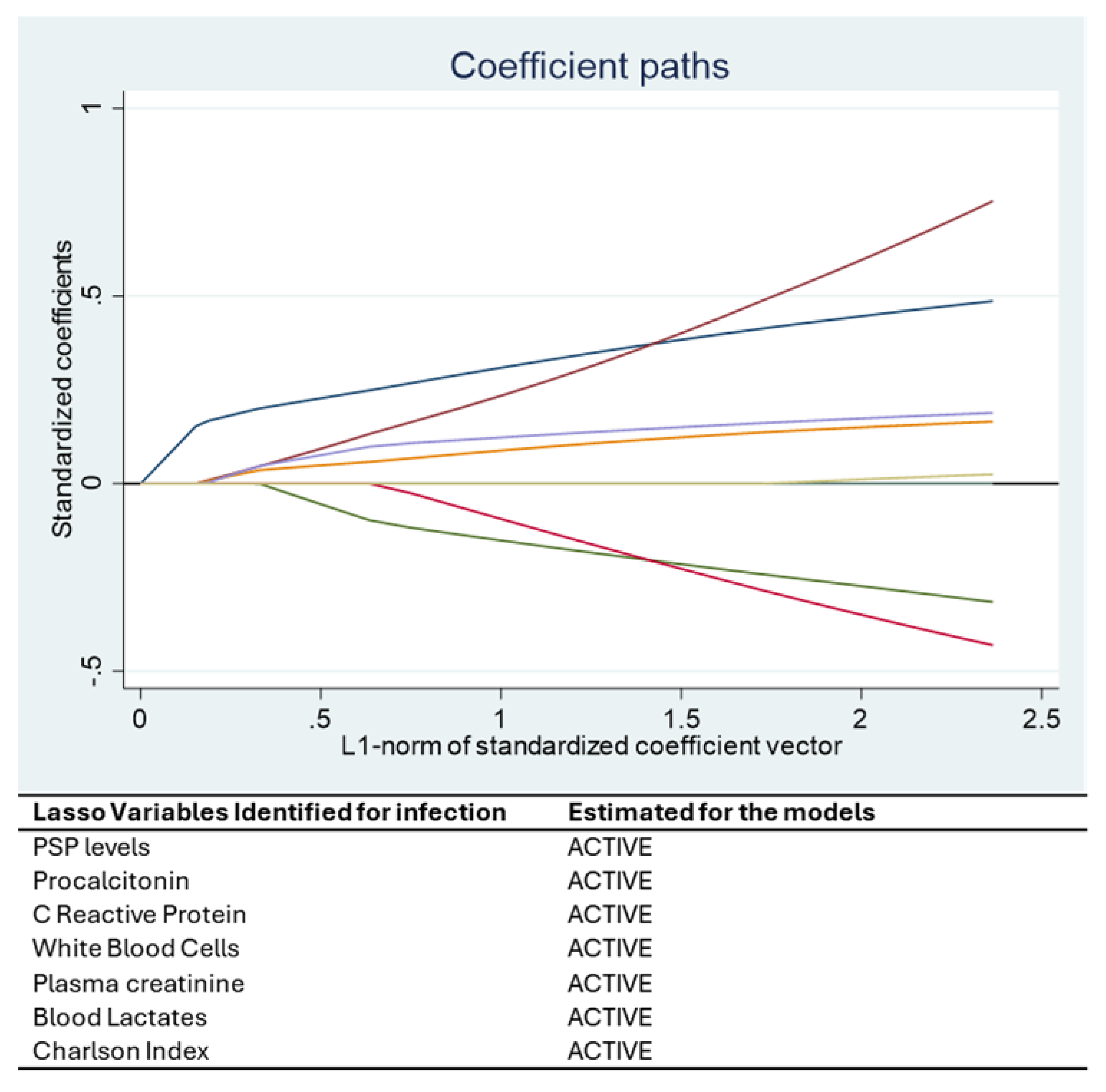
Statistical analysis was performed using STATA 16® (StataCorp, College Station, TX, USA). Continuous variables were presented as mean ± standard deviation or median [IQR], and categorical variables as percentages. A Least Absolute Shrinkage and Selection Operator (LASSO) regression was applied to identify the most relevant predictors of bacterial infection. This machine learning approach addresses multicollinearity and selects variables that optimize model performance.
The optimal value of the penalty parameter (λ) in the LASSO regression was automatically selected using 10-fold cross-validation, as implemented in STATA’s LASSO command. This approach balances model complexity and predictive performance by minimizing the cross-validated mean squared error [
12,
13,
14]. Probit models were used for binary outcomes, and linear regression for continuous variables. To account for repeated measurements over time and control for within-subject variability, we used mixed-effects models (random intercept models). These models allowed for simultaneous evaluation of associations between biomarker levels and infection (or severe respiratory failure) while adjusting for time effects and inter-individual differences. The fixed-effect variables included in the mixed models were those selected by the LASSO regression, ensuring consistency and minimizing overfitting. Random effects accounted for patient-level variation. Model performance was evaluated using receiver operating characteristic (ROC) curves, with an area under the curve (AUC) ≥ 0.7 considered acceptable for diagnostic discrimination. Statistical significance was set at
p ≤ 0.05. Additional model fit was assessed via the Akaike Information Criterion (AIC) and the Bayesian Information Criterion (BIC). Post hoc power analysis was performed to assess the statistical power of the main findings.
4. Discussion
In the course of our study, all considered biomarkers were analyzed using advanced machine learning (ML) techniques. These methodologies, particularly the Least Absolute Shrinkage and Selection Operator (LASSO) and mixed-effects models, enable the development of predictive models capable of handling multicollinearity, selecting relevant predictors, and minimizing overfitting. ML, through Artificial Intelligence (AI), can process complex datasets and simulate human-like reasoning abilities in diagnostics and decision-making [
10]. One of its key strengths lies in its ability to reduce human biases, such as selection bias, by applying algorithmic rules that are not influenced by subjective judgments [
11,
12]. The use of data-driven approaches further supports this objective, allowing the identification of robust, statistically validated predictors through the systematic evaluation of available data [
13].
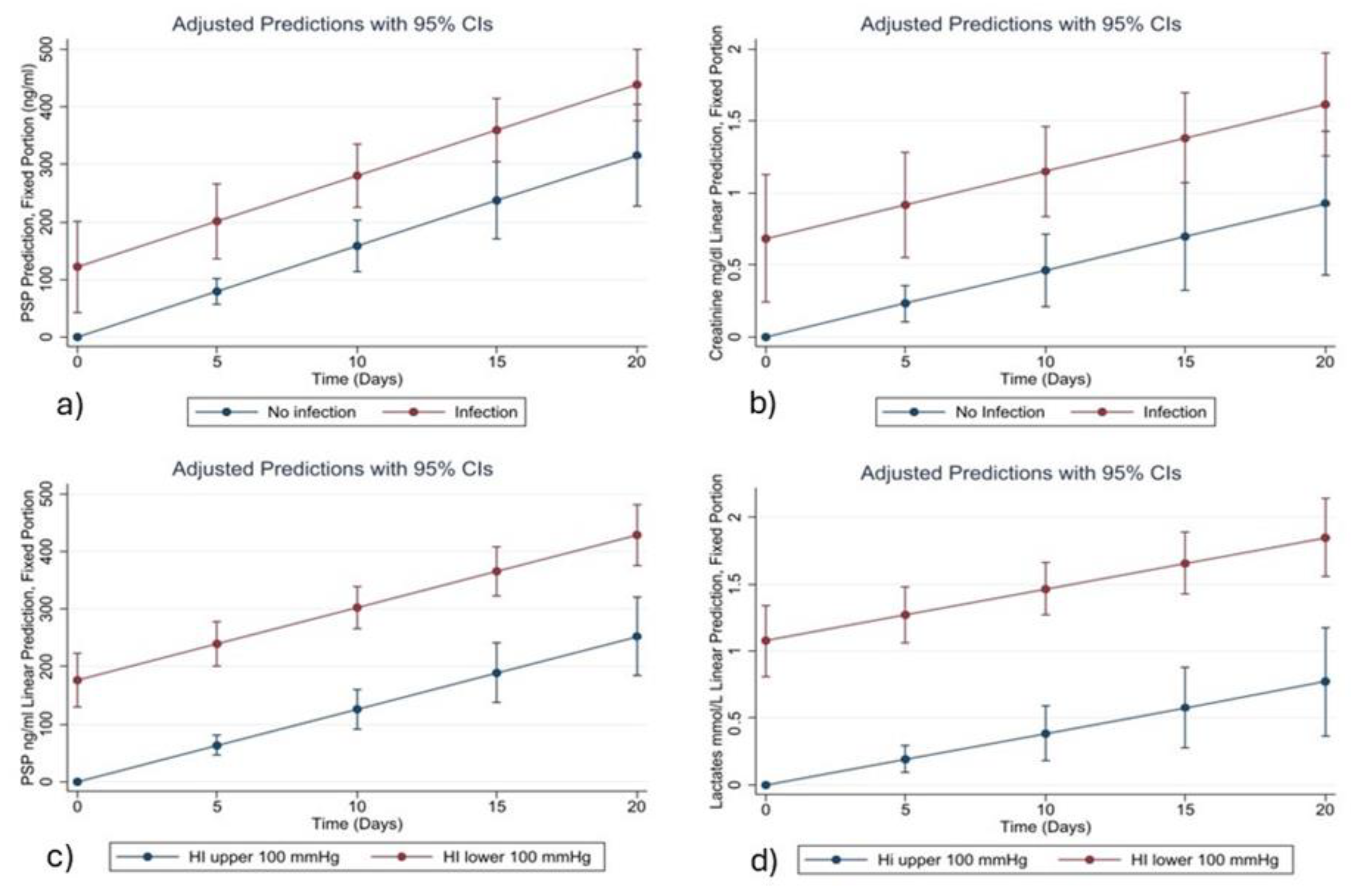
Among all the variables analyzed, Pancreatic Stone Protein (PSP) consistently emerged as a strong and independent biomarker for bacterial infection. The statistical relationships identified in our models are supported by clear biological mechanisms. PSP is secreted by the pancreas and various epithelial tissues in response to inflammatory and infectious stimuli. Its early and sustained rise in systemic infections is associated with neutrophil activation, increased endothelial permeability, and immune system mobilization. This biological behavior underpins its association with the onset of infection in critically ill COVID-19 patients, as observed in our study and in previous research [
3,
4].
Similarly, CRP and Procalcitonin (PCT) are well-known acute-phase reactants. CRP is synthesized by hepatocytes in response to IL-6, while PCT is released following bacterial endotoxin stimulation. Although both are widely used in clinical practice, their temporal trends in our cohort were less consistent than those of PSP, especially in the later phases of ICU stay. This may suggest that PSP has a more stable and linear correlation with infection severity and progression in this population.
Creatinine also emerged as a relevant variable in both the LASSO and mixed-effects models. While traditionally associated with renal function, its predictive value in our infection model likely reflects the high incidence of sepsis-associated acute kidney injury (AKI) in COVID-19 ICU patients. SARS-CoV-2 is known to invade tubular epithelial cells via ACE2 receptors, triggering direct cytopathic effects and inflammatory cascades leading to tubular necrosis [
15,
16,
17]. As such, elevated creatinine may serve as a proxy for systemic deterioration and infection-related organ dysfunction. While some studies have highlighted the superiority of eGFR over creatinine as a prognostic indicator [
18], others, including that of Malik et al., have confirmed its significant correlation with infection severity and mortality in COVID-19 [
19].
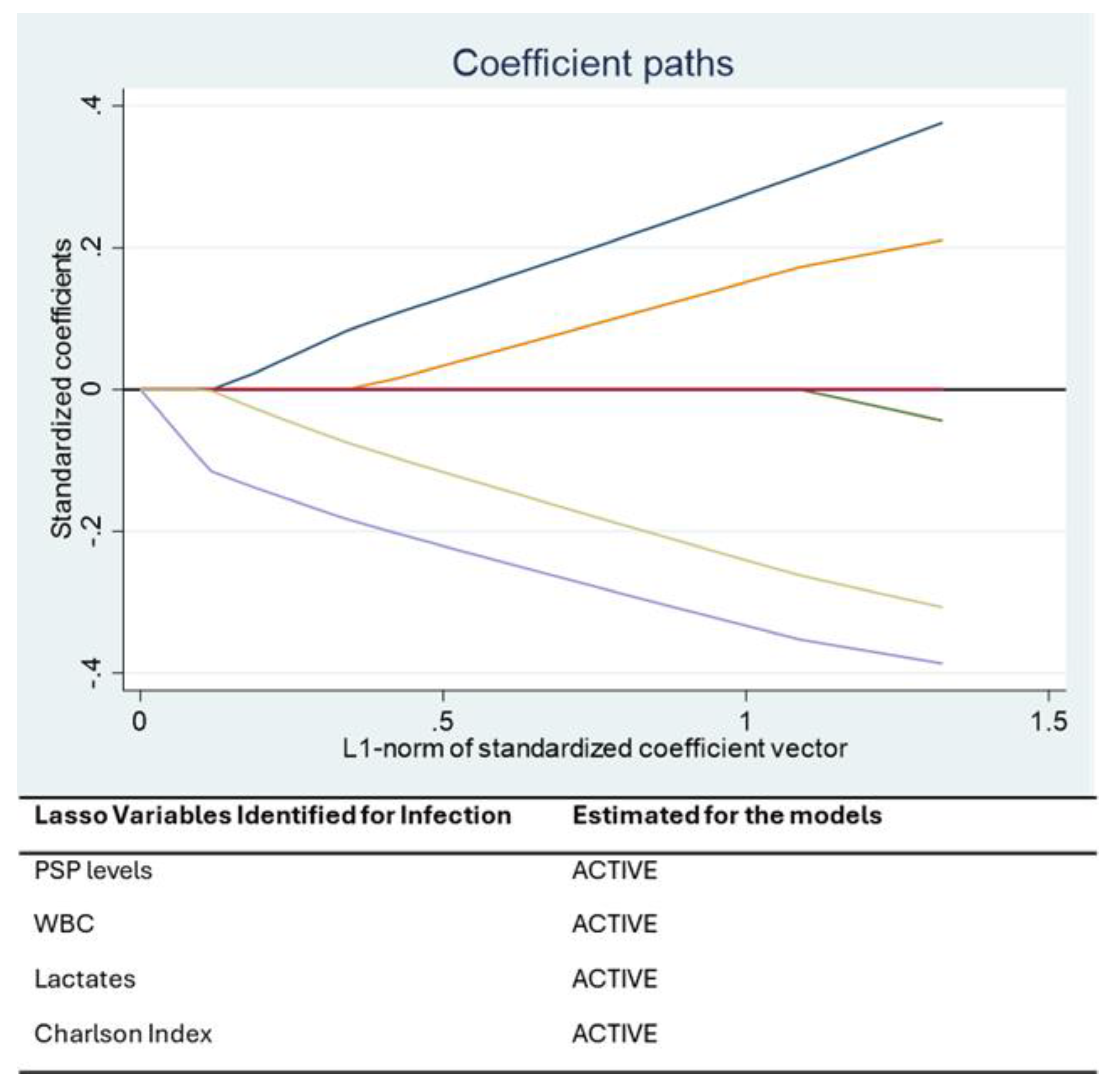
The prognostic value of PSP was also confirmed in the subgroup of patients experiencing severe respiratory failure, defined by a Horowitz Index (PaO2/FiO2) < 100 mmHg. In these patients, PSP, lactates, and Charlson Comorbidity Index were retained as significant predictors by the LASSO and mixed-effects models. The association between elevated PSP and respiratory failure may reflect systemic inflammation and the presence of pulmonary bacterial superinfection. Meanwhile, elevated lactate levels are indicative of impaired oxygen delivery and tissue hypoperfusion—both hallmark features of severe acute respiratory distress syndrome (ARDS). These biomarkers therefore provide complementary information and reflect different, yet interrelated, pathophysiological processes.
Our findings are consistent with results of previous studies. Lagadinou et al. demonstrated a correlation between elevated PSP levels and mortality in COVID-19 pneumonia [
14]. Van Singer et al. found that PSP, alone or in combination with the CURB-65 score, outperformed SOFA in predicting poor outcomes [
20]. Melegari et al. observed that PSP levels between 250 and 300 ng/mL at ICU admission were associated with a 50% risk of 90-day mortality in critically ill COVID-19 patients [
7]. This suggests that PSP could serve not only as a diagnostic but also a prognostic marker, warranting further investigation and potential integration into routine clinical workflows.
Yamada et al. also explored biomarkers of respiratory failure in COVID-19, identifying CRP, IL-6, D-dimer, and thrombocytopenia as significant markers [
21]. While our findings highlight a different set of predictors, particularly PSP and lactates, they complement the existing literature by expanding understanding of the pathophysiology underlying severe respiratory failure in SARS-CoV-2 infection.
Strengths and Limitations
The size of the sample in the present study was adequate with respect to statistical power, but the patient cohort was numerically limited. Therefore, it is necessary that larger cohorts be used for future, more in-depth studies. A post hoc power analysis was performed to assess the statistical power of the main findings. Based on the observed effect size in the probit model (Cohen’s d equivalent ≈ 0.5), the sample size of 130 patients, and a significance level (α) of 0.05, the calculated power exceeded 0.85 for detecting significant associations between selected predictors and the outcome of bacterial infection. This indicates that the study had an adequate sample size to detect clinically meaningful effects. The application of ML and a data-driven approach offers the advantage of a reduction in human bias, especially selection bias. A limitation of our approach is the lack of explicit control for certain potential confounders such as timing of antibiotic therapy or immunosuppressive treatments, which may influence biomarker dynamics. Potential confounders such as immunosuppressive treatments and timing of antibiotic therapy were considered clinically but were not included in the final mixed models due to multicollinearity concerns and limited sample size. LASSO-based variable selection aimed to minimize overfitting while optimizing predictive accuracy.
5. Conclusions
Our study confirms the potential role of Pancreatic Stone Protein (PSP) as a reliable biomarker for early identification and monitoring of bacterial infections in critically ill COVID-19 patients. Compared to traditional markers such as Procalcitonin and C-Reactive Protein, PSP showed a more consistent and linear association with infection and disease severity over time. The integration of PSP into predictive models based on machine learning, such as LASSO regression and mixed-effects models, highlighted its strong statistical and biological relevance.
Moreover, the identification of creatinine and lactate as complementary biomarkers supports the notion that multi-organ dysfunction, particularly involving the kidneys and perfusion status, significantly contributes to the risk of infection and respiratory failure in this population. The use of advanced statistical methods enabled a robust and unbiased selection of variables, further reinforcing the clinical applicability of our findings.
Although limited by the small sample size, the study offers promising insights that warrant further research. Larger multicenter studies are needed to validate PSP’s prognostic value and to explore its utility in guiding antibiotic stewardship strategies and improving risk stratification in intensive care settings.