EFCAB4B (CRACR2A/Rab46) Genetic Variants Associated with COVID-19 Fatality
Abstract
1. Introduction
2. Materials and Methods
2.1. Demographic Data
2.2. Analysis of EFCAB4B Variants Associated with COVID-19 Outcomes
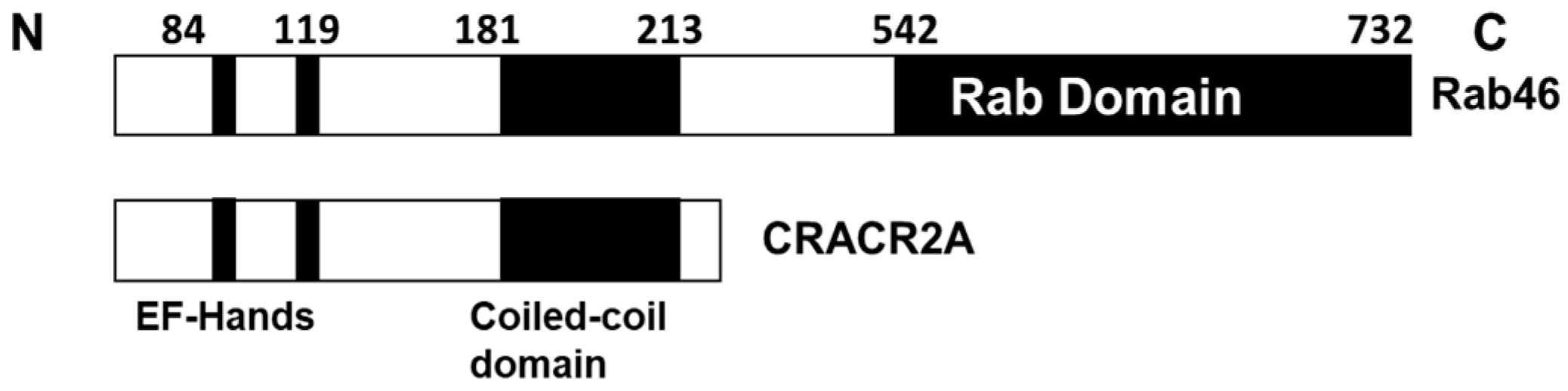
2.3. Conservation of Rab46 Protein Sequence across Different Species
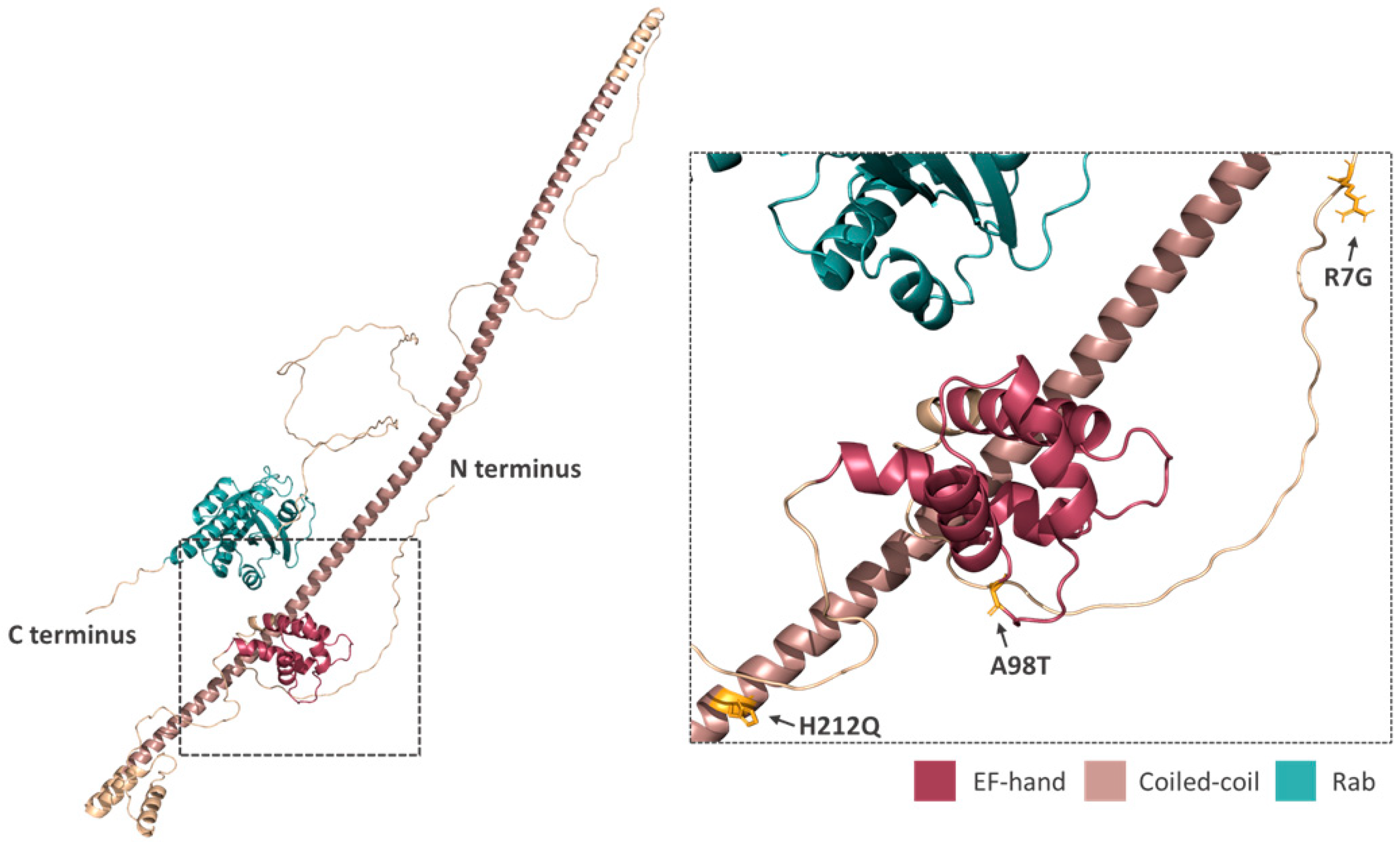
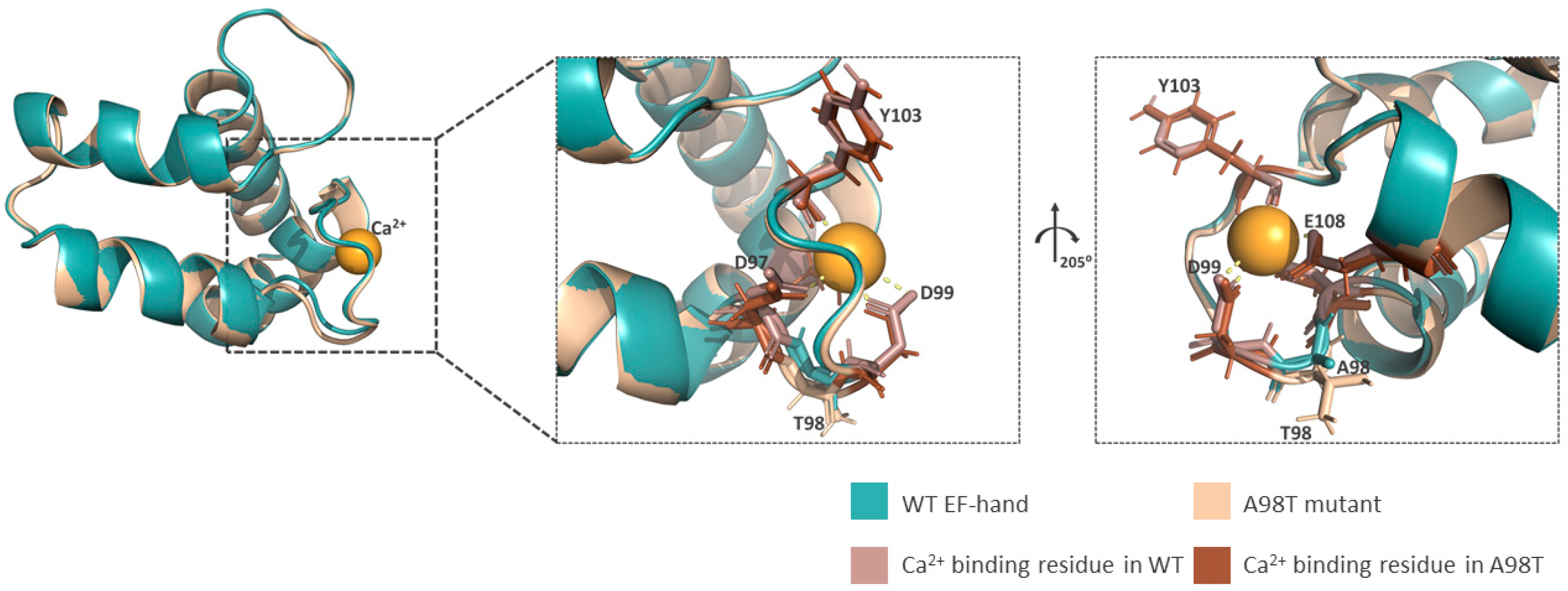
2.4. Rab46 Structural Analysis
3. Results
3.1. Demographics
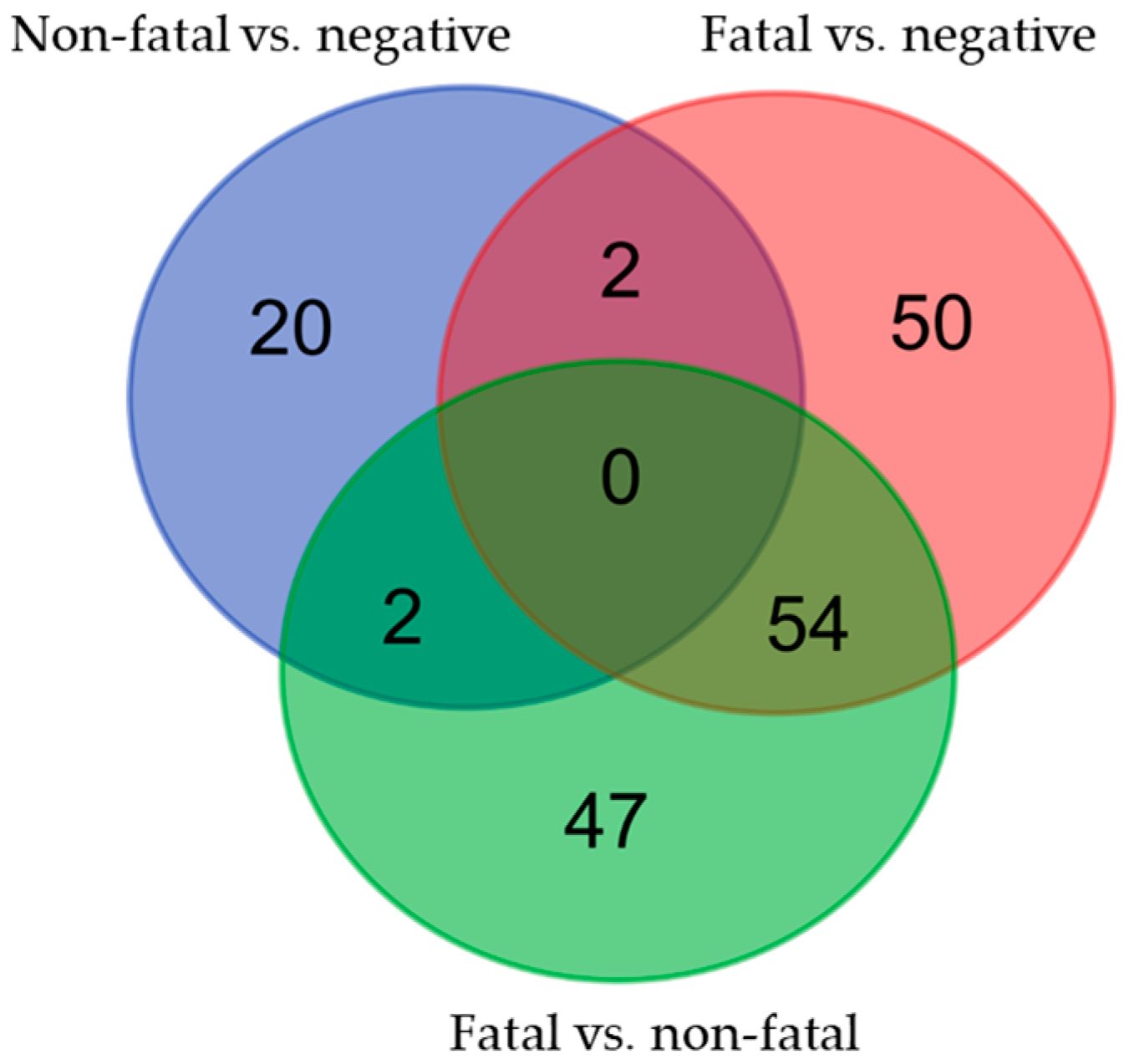
3.2. Variants in the EFCAB4B Gene Are Associated with Susceptibility to COVID-19 Infection and COVID-19 Fatality
3.3. EFCAB4B Missense Variants Associated with COVID-19
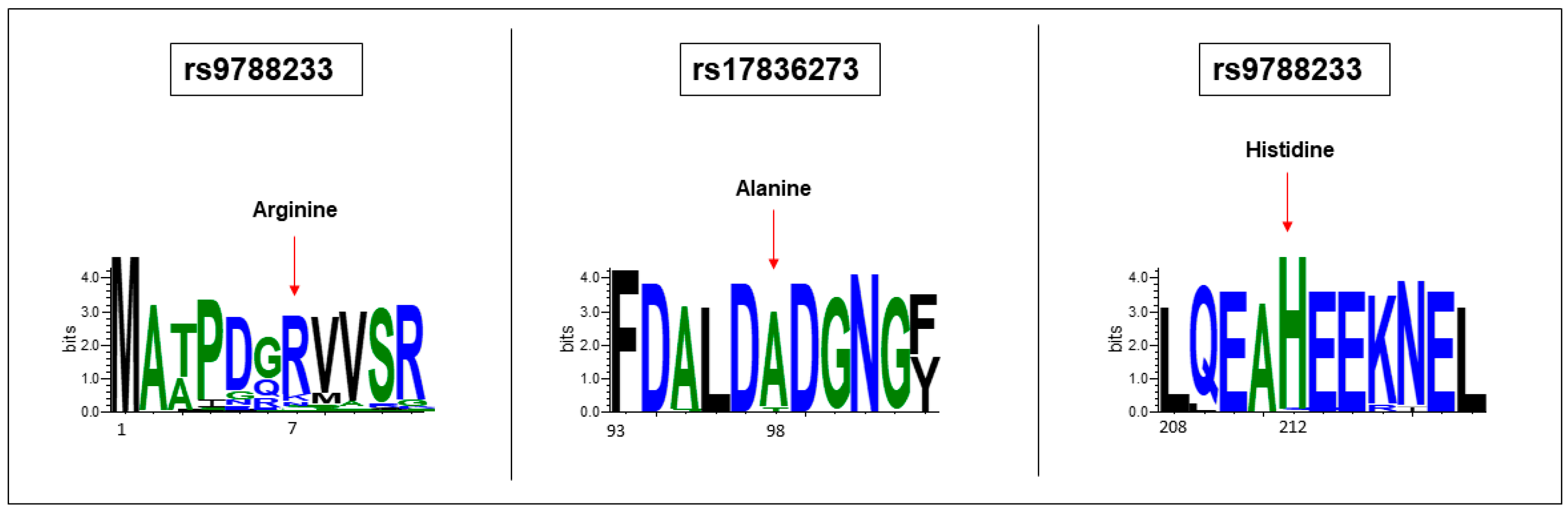
3.4. EFCAB4B Missense Variants Affect Highly Conserved Residues
3.5. Prediction of Missense Variants on Rab46 Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atzrodt, C.L.; Maknojia, I.; McCarthy, R.D.P.; Oldfield, T.M.; Po, J.; Ta, K.T.L.; Stepp, H.E.; Clements, T.P. A Guide to COVID-19: A global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020, 287, 3633–3650. [Google Scholar] [CrossRef]
- McFadyen, J.D.; Stevens, H.; Peter, K. The Emerging Threat of (Micro)Thrombosis in COVID-19 and Its Therapeutic Implications. Circ. Res. 2020, 127, 571–587. [Google Scholar] [CrossRef]
- Wazny, V.; Siau, A.; Wu, K.X.; Cheung, C. Vascular underpinning of COVID-19. Open Biol. 2020, 10, 200208. [Google Scholar] [CrossRef]
- Nägele, M.P.; Haubner, B.; Tanner, F.C.; Ruschitzka, F.; Flammer, A.J. Endothelial dysfunction in COVID-19: Current findings and therapeutic implications. Atherosclerosis 2020, 314, 58–62. [Google Scholar] [CrossRef]
- Zhang, J.; Tecson, K.M.; McCullough, P.A. Endothelial dysfunction contributes to COVID-19-associated vascular inflammation and coagulopathy. Rev. Cardiovasc. Med. 2020, 21, 315–319. [Google Scholar] [CrossRef]
- Goshua, G.; Pine, A.B.; Meizlish, M.L.; Chang, C.H.; Zhang, H.; Bahel, P.; Baluha, A.; Bar, N.; Bona, R.D.; Burns, A.J.; et al. Endotheliopathy in COVID-19-associated coagulopathy: Evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020, 7, e575–e582. [Google Scholar] [CrossRef]
- Villa, E.; Critelli, R.; Lasagni, S.; Melegari, A.; Curatolo, A.; Celsa, C.; Romagnoli, D.; Melegari, G.; Pivetti, A.; Di Marco, L.; et al. Dynamic angiopoietin-2 assessment predicts survival and chronic course in hospitalized patients with COVID-19. Blood Adv. 2021, 5, 662–673. [Google Scholar] [CrossRef]
- Fenyves, B.G.; Mehta, A.; Kays, K.R.; Beakes, C.; Margolin, J.; Goldberg, M.B.; Hacohen, N.; Filbin, M.R. Plasma P-selectin is an early marker of thromboembolism in COVID-19. Am. J. Hematol. 2021, 96, E468–E471. [Google Scholar] [CrossRef]
- Yatim, N.; Boussier, J.; Chocron, R.; Hadjadj, J.; Philippe, A.; Gendron, N.; Barnabei, L.; Charbit, B.; Szwebel, T.A.; Carlier, N.; et al. Platelet activation in critically ill COVID-19 patients. Ann. Intensive Care 2021, 11, 113. [Google Scholar] [CrossRef]
- Cotter, A.H.; Yang, S.T.; Shafi, H.; Cotter, T.M.; Palmer-Toy, D.E. Elevated von Willebrand Factor Antigen Is an Early Predictor of Mortality and Prolonged Length of Stay for Coronavirus Disease 2019 (COVID-19) Inpatients. Arch. Pathol. Lab. Med. 2022, 146, 34–37. [Google Scholar] [CrossRef]
- Philippe, A.; Chocron, R.; Gendron, N.; Bory, O.; Beauvais, A.; Peron, N.; Khider, L.; Guerin, C.L.; Goudot, G.; Levasseur, F.; et al. Circulating Von Willebrand factor and high molecular weight multimers as markers of endothelial injury predict COVID-19 in-hospital mortality. Angiogenesis 2021, 24, 505–517. [Google Scholar] [CrossRef]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef]
- Naß, J.; Terglane, J.; Gerke, V. Weibel Palade Bodies: Unique Secretory Organelles of Endothelial Cells that Control Blood Vessel Homeostasis. Front. Cell Dev. Biol. 2021, 9, 813995. [Google Scholar] [CrossRef]
- Karampini, E.; Bierings, R.; Voorberg, J. Orchestration of Primary Hemostasis by Platelet and Endothelial Lysosome-Related Organelles. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1441–1453. [Google Scholar] [CrossRef]
- Kawecki, C.; Lenting, P.J.; Denis, C.V. von Willebrand factor and inflammation. J. Thromb. Haemost. 2017, 15, 1285–1294. [Google Scholar] [CrossRef]
- Cossutta, M.; Darche, M.; Carpentier, G.; Houppe, C.; Ponzo, M.; Raineri, F.; Vallée, B.; Gilles, M.E.; Villain, D.; Picard, E.; et al. Weibel-Palade Bodies Orchestrate Pericytes During Angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1843–1858. [Google Scholar] [CrossRef]
- Wilson, L.A.; McKeown, L.; Tumova, S.; Li, J.; Beech, D.J. Expression of a long variant of CRACR2A that belongs to the Rab GTPase protein family in endothelial cells. Biochem. Biophys. Res. Commun. 2015, 456, 398–402. [Google Scholar] [CrossRef]
- Miteva, K.T.; Pedicini, L.; Wilson, L.A.; Jayasinghe, I.; Slip, R.G.; Marszalek, K.; Gaunt, H.J.; Bartoli, F.; Deivasigamani, S.; Sobradillo, D.; et al. Rab46 integrates Ca(2+) and histamine signaling to regulate selective cargo release from Weibel-Palade bodies. J. Cell Biol. 2019, 218, 2232–2246. [Google Scholar] [CrossRef]
- Srikanth, S.; Kim, K.D.; Gao, Y.; Woo, J.S.; Ghosh, S.; Calmettes, G.; Paz, A.; Abramson, J.; Jiang, M.; Gwack, Y. A large Rab GTPase encoded by CRACR2A is a component of subsynaptic vesicles that transmit T cell activation signals. Sci. Signal 2016, 9, ra31. [Google Scholar] [CrossRef]
- Wu, B.; Rice, L.; Shrimpton, J.; Lawless, D.; Walker, K.; Carter, C.; McKeown, L.; Anwar, R.; Doody, G.M.; Srikanth, S.; et al. Biallelic mutations in calcium release activated channel regulator 2A (CRACR2A) cause a primary immunodeficiency disorder. Elife 2021, 10, e72559. [Google Scholar] [CrossRef]
- Srikanth, S.; Jung, H.J.; Kim, K.D.; Souda, P.; Whitelegge, J.; Gwack, Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 2010, 12, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.S.; Srikanth, S.; Kim, K.D.; Elsaesser, H.; Lu, J.; Pellegrini, M.; Brooks, D.G.; Sun, Z.; Gwack, Y. CRACR2A-Mediated TCR Signaling Promotes Local Effector Th1 and Th17 Responses. J. Immunol. 2018, 201, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Talla, A.; Vasaikar, S.V.; Lemos, M.P.; Moodie, Z.; Lee Pebworth, M.-P.; Henderson, K.E.; Cohen, K.W.; Czartoski, J.L.; Lai, L.; Suthar, M.S.; et al. Longitudinal immune dynamics of mild COVID-19 define signatures of recovery and persistence. bioRxiv 2021. [Google Scholar] [CrossRef]
- Patel, M.A.; Knauer, M.J.; Nicholson, M.; Daley, M.; Van Nynatten, L.R.; Cepinskas, G.; Fraser, D.D. Organ and cell-specific biomarkers of Long-COVID identified with targeted proteomics and machine learning. Mol. Med. 2023, 29, 26. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Kosmicki, J.A.; Horowitz, J.E.; Banerjee, N.; Lanche, R.; Marcketta, A.; Maxwell, E.; Bai, X.; Sun, D.; Backman, J.D.; Sharma, D.; et al. A catalog of associations between rare coding variants and COVID-19 outcomes. medRxiv. 2021. [Google Scholar] [CrossRef]
- Horowitz, J.E.; Kosmicki, J.A.; Damask, A.; Sharma, D.; Roberts, G.H.L.; Justice, A.E.; Banerjee, N.; Coignet, M.V.; Yadav, A.; Leader, J.B.; et al. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat. Genet. 2022, 54, 382–392. [Google Scholar] [CrossRef]
- Lee, I.G.; Cason, S.E.; Alqassim, S.S.; Holzbaur, E.L.F.; Dominguez, R. A tunable LIC1-adaptor interaction modulates dynein activity in a cargo-specific manner. Nat. Commun. 2020, 11, 5695. [Google Scholar] [CrossRef]
- Boyle, A.L.; Rabe, M.; Crone, N.S.A.; Rhys, G.G.; Soler, N.; Voskamp, P.; Pannu, N.S.; Kros, A. Selective coordination of three transition metal ions within a coiled-coil peptide scaffold. Chem. Sci. 2019, 10, 7456–7465. [Google Scholar] [CrossRef]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef]
- Misceo, D.; Holmgren, A.; Louch, W.E.; Holme, P.A.; Mizobuchi, M.; Morales, R.J.; De Paula, A.M.; Stray-Pedersen, A.; Lyle, R.; Dalhus, B.; et al. A dominant STIM1 mutation causes Stormorken syndrome. Hum. Mutat. 2014, 35, 556–564. [Google Scholar] [CrossRef]
- Noury, J.B.; Böhm, J.; Peche, G.A.; Guyant-Marechal, L.; Bedat-Millet, A.L.; Chiche, L.; Carlier, R.Y.; Malfatti, E.; Romero, N.B.; Stojkovic, T. Tubular aggregate myopathy with features of Stormorken disease due to a new STIM1 mutation. Neuromuscul. Disord. 2017, 27, 78–82. [Google Scholar] [CrossRef]
- Furuyama, W.; Shifflett, K.; Pinski, A.N.; Griffin, A.J.; Feldmann, F.; Okumura, A.; Gourdine, T.; Jankeel, A.; Lovaglio, J.; Hanley, P.W.; et al. Rapid protection from COVID-19 in nonhuman primates vaccinated intramuscularly but not intranasally with a single dose of a recombinant vaccine. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wu, B.; Ramaiah, A.; Garcia, G., Jr.; Gwack, Y.; Arumugaswami, V.; Srikanth, S. ORAI1 establishes resistance to SARS-CoV-2 infection by regulating tonic type I interferon signaling. bioRxiv 2021. [Google Scholar] [CrossRef]
- Srikanth, S.; Woo, J.S.; Wu, B.; El-Sherbiny, Y.M.; Leung, J.; Chupradit, K.; Rice, L.; Seo, G.J.; Calmettes, G.; Ramakrishna, C.; et al. The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 2019, 20, 152–162. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Matuozzo, D.; Talouarn, E.; Marchal, A.; Zhang, P.; Manry, J.; Seeleuthner, Y.; Zhang, Y.; Bolze, A.; Chaldebas, M.; Milisavljevic, B.; et al. Rare predicted loss-of-function variants of type I IFN immunity genes are associated with life-threatening COVID-19. Genome Med. 2023, 15, 22. [Google Scholar] [CrossRef]

| COVID-19 Positive | ||||||
|---|---|---|---|---|---|---|
| Characteristic | Total N | COVID-19 Negative | Fatal and Non-Fatal | Fatal | Non-Fatal | p-Value |
| Number of Individuals | 10,118 | 8853 | 1265 | 194 | 1071 | |
| Age | 70 ± 8.3 | 70.1 ± 8.2 | 68.9 ± 9.2 | 75.1 ± 5.7 | 67.7 ± 9.2 | 1.38 × 10−30 |
| Sex | ||||||
| Male | 4972 | 4309 | 663 | 129 | 534 | 5.09 × 10−6 |
| Female | 5146 | 4544 | 602 | 65 | 537 | |
| Co-Morbidity | ||||||
| Myocardial Infarction | 429 | 375 | 54 | 13 | 41 | 0.19 |
| Stroke | 256 | 221 | 35 | 6 | 29 | 0.81 |
| Arthritis | 1477 | 1325 | 152 | 37 | 115 | 2.16 × 10−4 |
| Diabetes Mellitus | 731 | 627 | 104 | 27 | 77 | 1.34 × 10−3 |
| Hypertension | 3436 | 3010 | 426 | 86 | 340 | 2.95 × 10−3 |
| Asthma | 1380 | 1222 | 158 | 17 | 141 | 0.12 |
| SNP ID | MAF (ALFA) | REF | ALT | Type | Amino Acid Change |
|---|---|---|---|---|---|
| rs9788233 | 0.119 | T | C | substitution | R7G |
| rs17836273 | 0.124 | C | T | substitution | A98T |
| rs36030417 | 0.145 | A | T | substitution | H212Q |
| SNP ID | Effect Allele | Non-Fatal vs. Negative p-Value/OR | Fatal vs. Negative p-Value/OR | Fatal vs. Non-Fatal p-Value/OR |
|---|---|---|---|---|
| rs9788233 | C | - | 0.004/1.511 | 0.021/1.487 |
| rs17836273 | T | - | 0.012/1.433 | 0.099/1.314 |
| rs36030417 | T | - | 0.013/1.393 | 0.054/1.351 |
| Patient Cohort | SNP ID | Effect Allele | Hypertension p-Value/OR | Diabetes p-Value/OR | Asthma p-Value/OR | Arthritis p-Value/OR | Stroke p-Value/OR | Myocardial Infarction p-Value/OR |
|---|---|---|---|---|---|---|---|---|
| Fatal vs. negative | rs9788233 | C | 0.004/1.512 | 0.004/1.516 | 0.005/1.505 | 0.004/1.51 | 0.004/1.511 | 0.004/1.511 |
| rs17836273 | T | 0.012/1.435 | 0.011/1.44 | 0.014/1.426 | 0.013/1.433 | 0.012/1.433 | 0.012/1.433 | |
| rs36030417 | T | 0.013/1.395 | 0.012/1.398 | 0.015/1.386 | 0.013/1.394 | 0.013/1.393 | 0.013/1.394 | |
| Fatal vs. non-fatal | rs9788233 | C | 0.022/1.481 | 0.018/1.498 | 0.022/1.482 | 0.024/1.474 | 0.022/1.483 | 0.021/1.487 |
| rs17836273 | T | 0.105/1.309 | 0.089/1.325 | 0.102/1.311 | 0.117/1.297 | 0.104/1.309 | 0.1/1.313 | |
| rs36030417 | T | 0.054/1.352 | 0.05/1.359 | 0.056/1.35 | 0.062/1.34 | 0.053/1.353 | 0.055/1.351 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Wiktor, S.D.; Cheng, C.W.; Simmons, K.J.; Money, A.; Pedicini, L.; Carlton, A.; Breeze, A.L.; McKeown, L. EFCAB4B (CRACR2A/Rab46) Genetic Variants Associated with COVID-19 Fatality. COVID 2024, 4, 1087-1099. https://doi.org/10.3390/covid4070075
Wang D, Wiktor SD, Cheng CW, Simmons KJ, Money A, Pedicini L, Carlton A, Breeze AL, McKeown L. EFCAB4B (CRACR2A/Rab46) Genetic Variants Associated with COVID-19 Fatality. COVID. 2024; 4(7):1087-1099. https://doi.org/10.3390/covid4070075
Chicago/Turabian StyleWang, Dapeng, Sabina D. Wiktor, Chew W. Cheng, Katie J. Simmons, Ashley Money, Lucia Pedicini, Asya Carlton, Alexander L. Breeze, and Lynn McKeown. 2024. "EFCAB4B (CRACR2A/Rab46) Genetic Variants Associated with COVID-19 Fatality" COVID 4, no. 7: 1087-1099. https://doi.org/10.3390/covid4070075
APA StyleWang, D., Wiktor, S. D., Cheng, C. W., Simmons, K. J., Money, A., Pedicini, L., Carlton, A., Breeze, A. L., & McKeown, L. (2024). EFCAB4B (CRACR2A/Rab46) Genetic Variants Associated with COVID-19 Fatality. COVID, 4(7), 1087-1099. https://doi.org/10.3390/covid4070075







