The Analyses of High Infectivity Mechanism of SARS-CoV-2 and Its Variants
Abstract
:1. Introduction
2. Methods
3. Results and Discussion
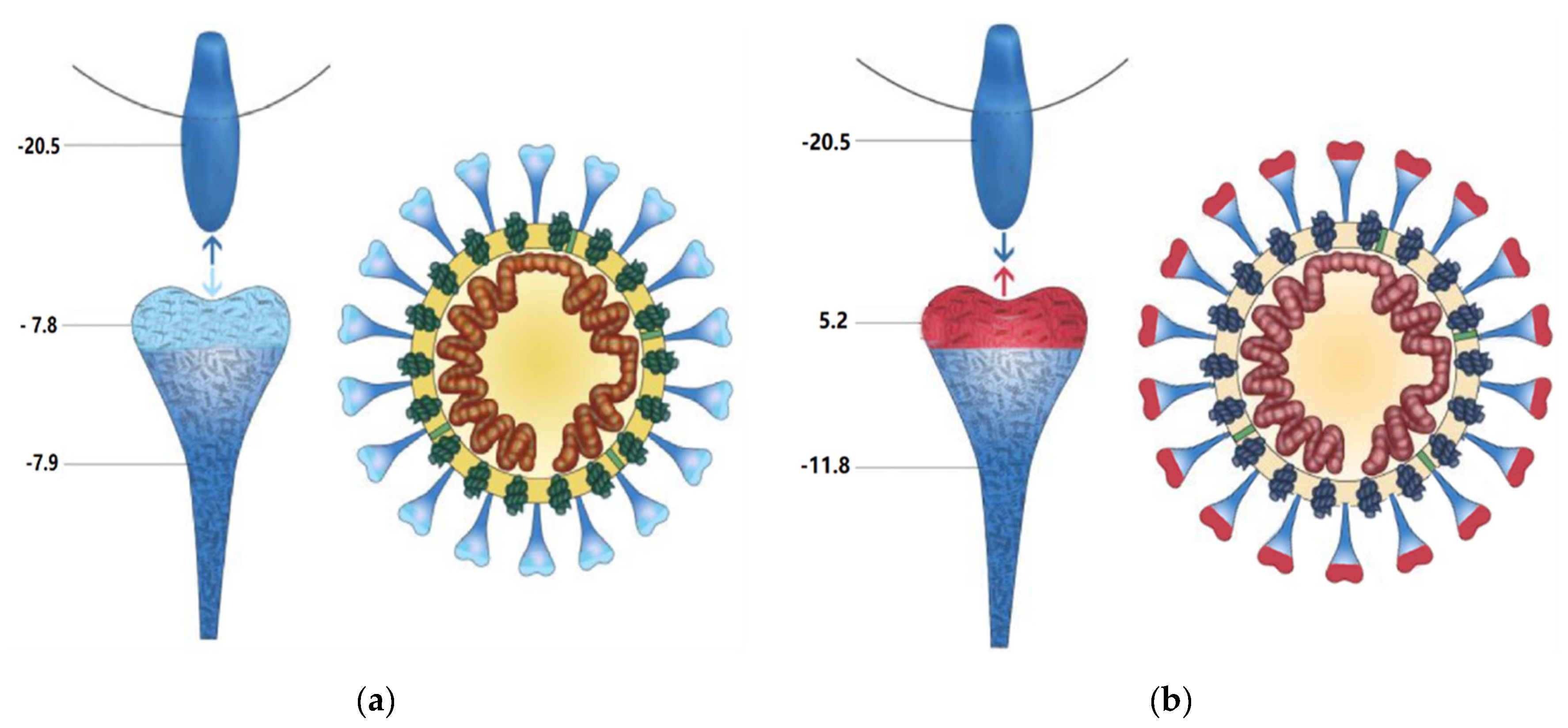
3.1. The Charge Distributions of SARS-CoV-2, SARS-CoV, and ACE2
3.2. Analysis of the SARS-CoV-2 Variants
3.3. Analysis of SARS-CoV-2 and SARS-CoV Antibodies
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chan, J.F.; Lau, S.K.; To, K.K.; Cheng, V.C.; Woo, P.C.; Yuen, K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Layne, S.P.; Hyman, J.M.; Morens, D.M.; Taubenberger, J.K. New coronavirus outbreak: Framing questions for pandemic prevention. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferretti, L.; Wymant, C.; Kendall, M.; Zhao, L.; Nurtay, A. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 2020, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, C.O.; West, A.P., Jr.; Huey-Tubman, K.E.; Hoffmann, M.A.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842.e16. [Google Scholar] [CrossRef] [PubMed]
- Zeyaullah, M.; AlShahrani, A.M.; Muzammil, K.; Ahmad, I.; Alam, S.; Khan, W.H.; Ahmad, R. COVID-19 and SARS-CoV-2 Variants: Current Challenges and Health Concern. Front Genet. 2021, 12, 693916. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Samad, A.B.A.; Amy, K.S. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19). Available online: https://www.ncbi.nlm.nih.gov/books/NBK570580/ (accessed on 18 July 2021.).
- Winger, A.; Caspari, T. The Spike of Concern-The Novel Variants of SARS-CoV-2. Viruses 2021, 13, 1002. [Google Scholar] [CrossRef]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Tolksdorf, B.; Nie, C.; Niemeyer, D.; Röhrs, V.; Berg, J.; Lauster, D.; Adler, J.M.; Haag, R.; Trimpert, J.; Kaufer, B.; et al. SARS-CoV-2 Variants: A Synopsis of In Vitro Effificacy Data of Convalescent Plasma, Currently Marketed Vaccines, and Monoclonal Antibodies. Viruses 2021, 13, 1211. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A potential therapeutic target for COVID-19. iScience 2020, 23, 101642. [Google Scholar] [CrossRef] [PubMed]
- Jaimes, J.A.; André, N.M.; Millet, J.K.; Whittaker, G.R. Structural modeling of 2019-novel coronavirus (nCoV) spike protein reveals a proteolytically sensitive activation loop as a distinguishing feature compared to SARS-CoV and related SARS-like coronaviruses. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.-H.; et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, T.T.-Y.; Jia, N.; Zhang, Y.-W.; Shum, M.H.-H.; Jiang, J.-F.; Zhu, H.-C.; Tong, Y.-G.; Shi, Y.-X.; Ni, X.-B.; Liao, Y.-S.; et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [Green Version]
- Kraemer, M.U.G.; Yang, C.-H.; Gutierrez, B.; Wu, C.-H.; Klein, B.; Pigott, D.M.; du Plessis, L.; Faria, N.R.; Li, R.; et al.; Open COVID-19 Data Working Group The effect of human mobility and control measures on the COVID-19 epidemic in China. Science 2020, 368, 493–497. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Yang, L.; Liu, R.; Liu, F.; Wu, K.-L.; Li, J.; Liu, X.-H.; Zhu, C.-L. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin. Chem. Lab. Med. 2020, 58, 1116–1120. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Lu, H.; Zhang, W. Clinical observation and management of COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 687–690. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boopathi, S.; Poma, A.B.; Kolandaivel, P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2021, 39, 3409–3418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.X.; Li, X.G.; Shu, C.B. A study of the number of charges of nucleic acid and protein. Chin. J. Org. Chem. 2000, 20, 401–406. [Google Scholar]
- Shen, T.; Wang, J.L.; Zhao, B.D. Biochemistry; Higher Education Press: Shanghai, China, 1980; Volume 1, pp. 132–134. [Google Scholar]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Volz, E.; Mishra, S.; Chand, M.; Barrett, J.C.; Johnson, R.; Geidelberg, L.; Hinsley, W.R.; Laydon, D.J.; Dabrera, G.; O’Toole, Á.; et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021, 593, 266–269. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv 2021. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Xia, H.; Zou, J.; Weaver, S.C.; Swanson, K.A.; Cai, H.; Cutler, M.; Cooper, D.; Muik, A.; et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 2021, 596, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S. Convergent Evolution of SARS-CoV-2 Spike Mutations, L452R, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lau, S.-Y.; Wang, P.; Mok, B.W.-Y.; Zhang, A.J.; Chu, H.; Lee, A.C.-Y.; Deng, S.; Chen, P.; Chan, K.-H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.W.; Toovey, O.T.; Harvey, K.N.; Hui, D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 2021, 82, e8–e10. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.K.; Dorosky, D.; Sharma, P.; Abbasi, S.A.; Dye, J.M.; Kranz, D.M.; Herbert, A.S.; Procko, E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom October to November 2020. Eur. Surveill. 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383.e9. [Google Scholar] [CrossRef]
- Wang, S.; Lu, Y.; Qian, W.H.; Zhao, Y.; Wang, Y.Y.; Zheng, S.Z.; Wang, A.Y. Cell charge: Potential important target of anti-tum or research. Chin. Pharmacol. Bull. 2010, 26, 1541–1544. [Google Scholar]
- Sarah, C.; David, T. SARS-CoV-2-induced remission of Hodgkin lymphoma. Br. J. Haematol. 2021, 192, 415. [Google Scholar] [CrossRef]
| Acidic Amino Acid | Kai | ΔKai | pKai | Basic Amino Acid | Kbi | ΔKbi | pKbi |
|---|---|---|---|---|---|---|---|
| Glu | 3.98 × 10−5 | 0 | 4.4 | Arg | 1.38 × 10−12 | 1.13 × 10−12 | 11.6–12.6 |
| Asp | 5.1 × 10−4 | 4.9 × 10−4 | 3.0–4.7 | Lys | 2.12 × 10−10 | 1.86 × 10−10 | 9.4–10.6 |
| - | - | - | - | His | 1.31 × 10−6 | 1.2 × 10−6 | 5.6–7.0 |
| Amino Acid | S1 | S2 | Sp | Ss1 | Ss2 | Ss | ACE2 |
|---|---|---|---|---|---|---|---|
| Glu | 23 | 25 | 48 | 17 | 25 | 42 | 56 |
| Asp | 31 | 31 | 62 | 45 | 28 | 73 | 43 |
| Arg | 29 | 13 | 42 | 23 | 16 | 39 | 31 |
| Lys | 30 | 31 | 61 | 31 | 29 | 60 | 47 |
| His | 9 | 8 | 17 | 9 | 6 | 15 | 16 |
| pH | 5.0 | 5.5 | 6.0 | 6.5 | 7.0 | 7.3 | 8.0 | 8.5 |
|---|---|---|---|---|---|---|---|---|
| HSp | 18.9 | 8.9 | 1.6 | −3.3 | −5.8 | −6.6 | −8.1 | −10.8 |
| ΔHSp | ±2.7 | ±3.6 | ±4.0 | ±2.5 | ±1.1 | ±0.8 | ±1.2 | ±3.2 |
| HSs | 7.1 | −1.9 | −8.3 | −12.7 | −14.9 | −15.6 | −17.1 | −19.7 |
| ΔHSs | ±2.7 | ±3.3 | ±3.5 | ±2.2 | ±1.0 | ±0.7 | ±1.2 | ±7.6 |
| HACE2 | 5.2 | −5.3 | −12.6 | −17.5 | −19.8 | −20.5 | −21.8 | −23.9 |
| ΔHACE2 | ±2.3 | ±3.3 | ±3.7 | ±2.3 | ±1.1 | ±0.7 | ±0.95 | ±2.5 |
| pH | 5.0 | 5.5 | 6.0 | 6.5 | 7.0 | 7.3 | 8.0 | 8.5 |
|---|---|---|---|---|---|---|---|---|
| Sp (RBD) | 8.5 | 7.3 | 6.6 | 6.3 | 6.1 | 6.0 | 5.8 | 5.2 |
| S1 | 18.2 | 13.2 | 9.5 | 6.9 | 5.6 | 5.2 | 4.4 | 3.1 |
| S2 | 0.7 | −4.3 | −7.9 | −10.2 | −11.4 | −11.8 | −12.6 | −13.9 |
| Ss (RBD) | 4.9 | 4.1 | 3.5 | 3.2 | 3.1 | 4.0 | 2.8 | 2.2 |
| Ss1 | 4.2 | −0.1 | −3.6 | −6.1 | −7.4 | −7.8 | −8.6 | −9.9 |
| Ss2 | 2.9 | −1.7 | −4.7 | −6.6 | −7.6 | −7.9 | −8.5 | −9.8 |
| SARS-CoV-2 Variant | SRBD Mutations | S1 Mutations Except RBD | S2 Mutations | Charges of Sp(RBD) at PH 7.3 | Charges of S1 at PH 7.3 | WHO Label | Other |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | - | - | - | 6.0 | 5.2 | - | - |
| Alpha (B.1.1.7) | N501Y | ∆H69, ΔV70, ΔY144, A570D, D614G, P681H | T716I S982A D1118H | 6.0 | 5.2 + 1 * | VOC | D1118H mutation decreased the Coulomb repulsion between S1 and ACE2 |
| B.1.1.7 + E484K | E484K N501Y | ∆H69, ΔV70, ΔY144, A570D, D614G, P681H | T716I S982A D1118H | 8.0 | 7.2 + 1 * | VOC | D1118H mutation decreased the Coulomb repulsion between S1 and ACE2 |
| Beta (B.1.351) | K417N, E484K, N501Y | L18F, D80A, D215G, R246I ∆L242, ∆A243, ∆L244, D614G | A701V | 7.0 | 8.2 | VOC | |
| Gamma (P.1) | K417T, E484K, N501Y | L18F, T20N, P26S, D138Y, R190S, H655Y, D614G | T1027I | 7.0 | 7.2 | VOC | |
| Delta (B.1.617.2) | L452R, T478K | T19R, ΔF157, ΔR158, D614G, P681R | D950N | 8.0 | 9.2 + 1 * | VOC | D950N mutation decreased the Coulomb repulsion between S1 and ACE2 |
| Delta Plus | K417N, L452R, T478K | T19R, ΔF157, ΔR158, D614G, P681R | D950N | 7.0 | 8.2 + 1 * | ||
| Mu | R346K, E484K, N501Y | D614G, P681H | - | 8.0 | 8.3 | VOI | |
| Lambda | L452Q, F490S | D614G | - | 6.0 | 6.2 | VOI |
| Net Charge | −7 | −6 | −5 | −4 | −3 | −2 | −1 | 0 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABR of SARS-CoV-2 | 4 | 3 | 5 | 3 | 6 | 15 | 8 | 9 | 5 | 1 | 0 | 2 |
| ABR of SARS-CoV | 0 | 0 | 0 | 2 | 7 | 8 | 14 | 17 | 9 | 3 | 0 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Zhao, T.; Lu, M.; Zhang, Y.; Yao, X.; Wu, G.; Dai, F.; Zhang, F.; Zhang, G. The Analyses of High Infectivity Mechanism of SARS-CoV-2 and Its Variants. COVID 2021, 1, 666-673. https://doi.org/10.3390/covid1040054
Lu Y, Zhao T, Lu M, Zhang Y, Yao X, Wu G, Dai F, Zhang F, Zhang G. The Analyses of High Infectivity Mechanism of SARS-CoV-2 and Its Variants. COVID. 2021; 1(4):666-673. https://doi.org/10.3390/covid1040054
Chicago/Turabian StyleLu, Yonghua, Tianfu Zhao, Ming Lu, Yaopeng Zhang, Xiang Yao, Guoyi Wu, Fangyin Dai, Fengxiu Zhang, and Guangxian Zhang. 2021. "The Analyses of High Infectivity Mechanism of SARS-CoV-2 and Its Variants" COVID 1, no. 4: 666-673. https://doi.org/10.3390/covid1040054
APA StyleLu, Y., Zhao, T., Lu, M., Zhang, Y., Yao, X., Wu, G., Dai, F., Zhang, F., & Zhang, G. (2021). The Analyses of High Infectivity Mechanism of SARS-CoV-2 and Its Variants. COVID, 1(4), 666-673. https://doi.org/10.3390/covid1040054







