Esterases: Mechanisms of Action, Biological Functions, and Application Prospects
Abstract
1. Introduction
2. Types and Sources of Esterases
3. Lipases
3.1. Sources of Lipases
3.2. Clinical Significance of Lipases
3.3. Industrial and Biotechnological Applications
4. Cutinases
4.1. Sources of Cutinases
4.2. Bacterial Cutinases
4.3. Plant Cutinases
4.4. Cutinase Applications
4.4.1. Degradation of Toxicants and Industrial Applications
4.4.2. Polyester Degradation
4.4.3. Medical Applications of Cutinases
5. Phospholipases
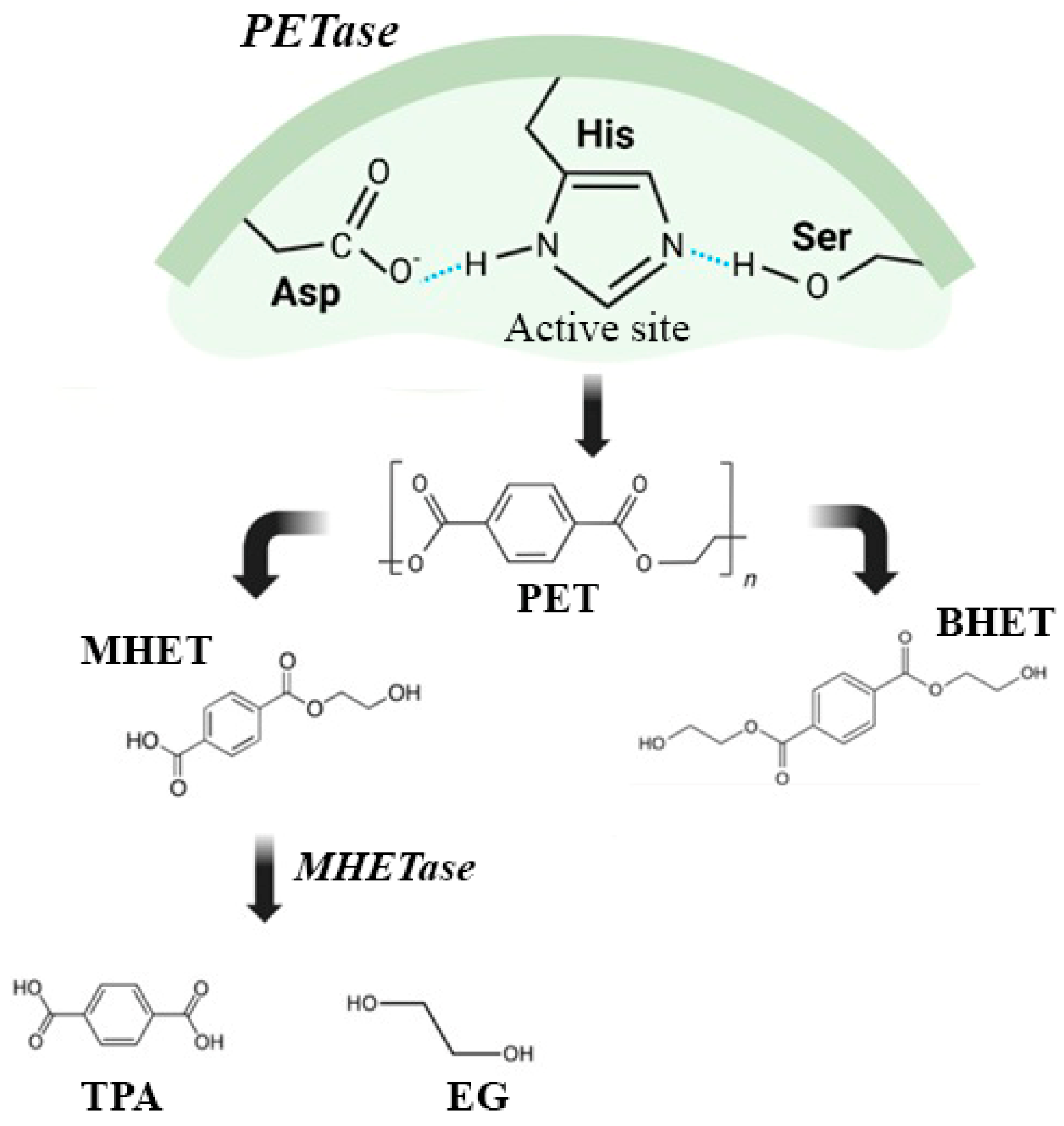
6. PETases
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EC | Enzyme classification |
| EG | Ethylene glycol |
| PCL | Polycaprolactone |
| PET | Polyethylene terephthalate |
| PETases | Polyethylene terephthalate hydrolases |
| PLA | Polylactic acid |
| TPA | Terephthalic acid |
References
- Barzkar, N.; Sohail, M.; Tamadoni Jahromi, S.; Gozari, M.; Poormozaffar, S.; Nahavandi, R.; Hafezieh, M. Marine bacterial esterases: Emerging biocatalysts for industrial applications. Appl. Biochem. Biotechnol. 2021, 193, 1187–1214. [Google Scholar] [CrossRef]
- De Luca, V.; Mandrich, L. Chapter 13—Lipases/esterases from extremophiles: Main features and potential biotechnological applications. In Physiological and Biotechnological Aspects of Extremophiles; Academic Press: Cambridge, MA, USA, 2020; pp. 169–181. [Google Scholar] [CrossRef]
- Tchigvintsev, A.; Tran, H.; Popovic, A.; Kovacic, F.; Brown, G.; Flick, R.; Hajighasemi, M.; Egorova, O.; Somody, J.C.; Tchigvintsev, D.; et al. The environment shapes microbial enzymes: Five cold-active and salt-resistant carboxylesterases from marine metagenomes. Appl. Microbiol. Biotechnol. 2015, 99, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Bessler, C.; Srinivas, R.; Krishna, S.H. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol. 2002, 20, 433–437. [Google Scholar] [CrossRef]
- Gupta, A.K.; Nayduch, D.; Verma, P.; Shah, B.; Ghate, H.V.; Patole, M.S.; Shouche, Y.S. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS Microbiol. Ecol. 2012, 79, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Ranjitha, P.; Karthy, E.S.; Mohankumar, A. Purification and characterization of the lipase from marine vibrio fischeri. Int. J. Biol. 2009, 1, 48–56. [Google Scholar] [CrossRef]
- Panda, T.; Gowrishankar, B.S. Production and applications of esterases. Appl. Microbiol. Biotechnol. 2005, 67, 160–169. [Google Scholar] [CrossRef]
- Aktayeva, S.; Khassenov, B. New Bacillus paralicheniformis strain with high proteolytic and keratinolytic activity. Sci. Rep. 2024, 14, 22621. [Google Scholar] [CrossRef]
- Aktayeva, S.; Khassenov, B. High keratinase and other types of hydrolase activity of the new strain of Bacillus paralicheniformis. PLoS ONE 2024, 19, e0312679. [Google Scholar] [CrossRef]
- Ufarté, L.; Laville, É.; Duquesne, S.; Potocki-Veronese, G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnol. Adv. 2015, 33, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Ramnath, L.; Sithole, B.; Govinden, R. Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can. J. Microbiol. 2017, 63, 179–192. [Google Scholar] [CrossRef]
- Hernández-Sánchez, B.; Díaz-Godínez, R.; Luna-Sánchez, S.; Sánchez, C. Esterase production by microorganisms: Importance and industrial application. Mex. J. Biotechnol. 2019, 4, 25–37. [Google Scholar] [CrossRef]
- Rehdorf, J.; Behrens, G.A.; Nguyen, G.-S.; Kourist, R.; Bornscheuer, U.T. Pseudomonas putida esterase contains a GGG(A)X-motif confering activity for the kinetic resolution of tertiary alcohols. Appl. Microbiol. Biotechnol. 2012, 93, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Kourist, R.; Jochens, H.; Bartsch, S.; Kuipers, R.; Padhi, S.K.; Gall, M.; Böttcher, D.; Joosten, H.J.; Bornscheuer, U.T. The α/β-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. ChemBioChem 2010, 11, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Lenfant, N.; Hotelier, T.; Velluet, E.; Bourne, Y.; Marchot, P.; Chatonnet, A. Esther, the database of the α/β-hydrolase fold superfamily of proteins: Tools to explore diversity of functions. Nucleic Acids Res. 2013, 41, D423–D429. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Cantu, D.C.; Chen, Y.; Lemons, M.L.; Reilly, P.J. ThYme: A database for thioester-active enzymes. Nucleic Acids Res. 2011, 39, D342–D346. [Google Scholar] [CrossRef]
- Chen, Y.; Black, D.S.; Reilly, P.J. Carboxylic ester hydrolases: Classification and database derived from their primary, secondary, and tertiary structures. Protein Sci. 2016, 25, 1942–1953. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Trono, D. Recombinant Lipases and Phospholipases and Their Use as Biocatalysts for Industrial Applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef]
- Casas-Godoy, L.; Gasteazoro, F.; Duquesne, S.; Bordes, F.; Marty, A.; Sandoval, G. Lipases: An Overview. Methods Mol. Biol. 2018, 1835, 3–38. [Google Scholar] [CrossRef]
- Moraleda-Muñoz, A.; Shimkets, L.J. Lipolytic enzymes in Myxococcus xanthus. J. Bacteriol. 2007, 189, 3072–3080. [Google Scholar] [CrossRef]
- Javed, S.; Azeem, F.; Hussain, S.; Rasul, I.; Siddique, M.H.; Riaz, M.; Afzal, M.; Kouser, A.; Nadeem, H.; Kouser, A.; et al. Bacterial lipases: A review on purification and characterization. Prog. Biophys. Mol. Biol. 2018, 132, 23–34. [Google Scholar] [CrossRef]
- Rajendran, A.; Palanisamy, A.; Thangavelu, V. Lipase catalyzed ester synthesis for food processing industries. Braz. Arch. Biol. Technol. 2009, 52, 207–219. [Google Scholar] [CrossRef]
- Karadzic, I.; Masui, A.; Zivkovic, L.I.; Fujiwara, N. Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metalworking fluid. J. Biosci. Bioeng. 2006, 102, 82–89. [Google Scholar] [CrossRef]
- Andualema, B.; Gessesse, A. Microbial lipases and their industrial applications: Review. Biotechnology 2012, 11, 100–118. [Google Scholar] [CrossRef]
- Prakash, D.; Nawani, N.; Prakash, M.; Bodas, M.; Mandal, A.; Khetmalas, M.; Kapadnis, B. Actinomycetes: A repertory of green catalysts with a potential revenue resource. BioMed Res. Int. 2013, 2013, 264020. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Carpen, A.; Bonomi, F.; Iametti, S.; Marengo, M. Effects of starch addition on the activity and specificity of food-grade lipases. Biotechnol. Appl. Biochem. 2019, 66, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Busk, P.K.; Lange, L. Characterization of a newsn-1,3-regioselective triacylglycerol lipase from Malbranchea cinnamomea. Biotechnol. Appl. Biochem. 2016, 63, 471–478. [Google Scholar] [CrossRef]
- Beatriz Vermelho, A.B.; Couri, S. Methods to Determine Enzymatic Activity; Bentham Science Publishers: Sharjah, United Arab Emirates, 2013. [Google Scholar] [CrossRef]
- Bracalente, F.; Sabatini, M.; Arabolaza, A.; Gramajo, H. Escherichia coli coculture for de novo production of esters derived of methyl-branched alcohols and multi-methyl branched fatty acids. Microb. Cell Fact. 2022, 21, 10. [Google Scholar] [CrossRef]
- De Maria, L.; Vind, J.; Oxenbøll, K.M.; Svendsen, A.; Patkar, S. Phospholipases and their industrial applications. Appl. Microbiol. Biotechnol. 2007, 74, 290–300. [Google Scholar] [CrossRef]
- Freitas, L.; Bueno, T.; Perez, V.H.; Santos, J.C.; de Castro, H.F. Enzymatic hydrolysis of soybean oil using lipase from different sources to yield concentrated of polyunsaturated fatty acids. World J. Microbiol. Biotechnol. 2007, 23, 1725–1731. [Google Scholar] [CrossRef]
- Bora, L.; Kalita, M.C. Production of thermostable alkaline lipase on vegetable oils from a thermophilic Bacillus sp. DH4, characterization and its potential applications as detergent additive. J. Chem. Technol. Biotechnol. 2008, 83, 688–693. [Google Scholar] [CrossRef]
- Adrio, J.L.; Demain, A.L. Microbial enzymes: Tools for biotechnological processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef]
- Terry, L.A.; White, S.F.; Tigwell, L.J. The application of biosensors to fresh produce and the wider food industry. J. Agric. Food Chem. 2005, 53, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Sarrouh, B. Up-to-date insight on industrial enzymes applications and global market. J. Bioprocess. Biotech. 2012, s4, 002. [Google Scholar] [CrossRef]
- Roohi, B.K.; Bano, K.; Kuddus, M.; Zaheer, M.R.; Zia, Q.; Khan, M.F.; Ashraf, G.M.; Gupta, A.; Aliev, G. Microbial enzymatic degradation of biodegradable plastics. Curr. Pharm. Biotechnol. 2017, 18, 429–440. [Google Scholar] [CrossRef]
- Vanleeuw, E.; Winderickx, S.; Thevissen, K.; Lagrain, B.; Dusselier, M.; Cammue, B.P.A.; Sels, B.F. Substrate-specificity of Candida rugosa lipase and its industrial application. ACS Sustain. Chem. Eng. 2019, 7, 15828–15844. [Google Scholar] [CrossRef]
- Badgujar, K.C.; Bhanage, B.M. Lipase immobilization on hyroxypropyl methyl cellulose support and its applications for chemo-selective synthesis of β-amino ester compounds. Process Biochem. 2016, 51, 1420–1433. [Google Scholar] [CrossRef]
- Sadaf, A.; Grewal, J.; Jain, I.; Kumari, A.; Khare, S.K. Stability and structure of Penicillium chrysogenum lipase in the presence of organic solvents. Prep. Biochem. Biotechnol. 2018, 48, 977–983. [Google Scholar] [CrossRef]
- Agboh, K.; Lau, C.H.F.; Khoo, Y.S.K.; Singh, H.; Raturi, S.; Nair, A.V.; Howard, J.; Chiapello, M.; Feret, R.; Deery, M.J.; et al. Powering the ABC multidrug exporter LmrA: How nucleotides embrace the ion-motive force. Sci. Adv. 2018, 4, eaas9365. [Google Scholar] [CrossRef] [PubMed]
- Walls, L.E.; Rios-Solis, L. Sustainable production of microbial isoprenoid derived advanced biojet fuels using different generation feedstocks: A review. Front. Bioeng. Biotechnol. 2020, 8, 599560. [Google Scholar] [CrossRef]
- Reetz, M.T. Biocatalysis in organic chemistry and biotechnology: Past, present, and future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef]
- Mendes, A.A.; Oliveira, P.C.; de Castro, H.F. Properties and biotechnological applications of porcine pancreatic lipase. J. Mol. Cat. B Enzym. 2012, 78, 119–134. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 169. [Google Scholar] [CrossRef]
- Dwivedi, S.K.; Enespa. In vitro cellulase activity of two wilt causing soil fusaria (Fusarium solani and F. oxysporum f. sp. lycopersici) and efficacy of some pesticides against the said fusaria. J. Appl. Hortic. 2015, 17, 58–65. [Google Scholar] [CrossRef]
- Salameh, M.; Wiegel, J. Lipases from extremophiles and potential for industrial applications. Adv. Appl. Microbiol. 2007, 61, 253–283. [Google Scholar] [CrossRef]
- Lipoprotein Lipase Enzyme Activity Assay Validation and Clinical Assessment. ClinicalTrials.gov ID NCT02656095. 11.07.2019. Available online: https://www.clinicaltrials.gov/study/NCT02656095?term=lipase&viewType=Table&rank=6&checkSpell= (accessed on 3 January 2023).
- Gangadhara, A.; Pasupala, P.; Radhakrishnan, S. Lipases: An overview of its current challenges and prospectives in the revolution of biocatalysis. Biocatal. Agr. Biotech. 2016, 7, 257–270. [Google Scholar] [CrossRef]
- Rani, G.; Arti, K.; Poonam, S.; Yogesh, S. Molecular and functional diversity of yeast and fungal lipases: Their role in biotechnology and cellular physiology. Prog. Lip. Res. 2015, 57, 40–54. [Google Scholar] [CrossRef]
- Liu, T.-T.; Liu, X.-T.; Chen, Q.-X.; Shi, Y. Lipase Inhibitors for Obesity: A Review. Biomed. Pharmacother. 2020, 128, 110314. [Google Scholar] [CrossRef] [PubMed]
- Papackova, Z.; Cahova, M. Fatty Acid Signaling: The New Function of Intracellular Lipases. Int. J. Mol. Sci. 2015, 16, 3831–3855. [Google Scholar] [CrossRef] [PubMed]
- Glybera Registry, Lipoprotein Lipase Deficient (LPLD) Patients (GENIALL). ClinicalTrials.gov ID NCT03293810. 27.11.2023. Available online: https://www.clinicaltrials.gov/study/NCT03293810?cond=%22Hyperlipoproteinemia%20Type%20I%22&viewType=Table&rank=1 (accessed on 3 January 2023).
- Acid Lipase Replacement Investigating Safety and Efficacy (ARISE) in Participants with Lysosomal Acid Lipase Deficiency (ARISE). ClinicalTrials.gov ID NCT01757184. 29.12.2020. Available online: https://clinicaltrials.gov/study/NCT01757184 (accessed on 3 January 2023).
- Identification of Undiagnosed Lysosomal Acid Lipase Deficiency. ClinicalTrials.gov ID NCT01716728. 13.08.2013. Available online: https://clinicaltrials.gov/study/NCT01716728?term=AREA%5BConditionSearch%5D(%22Wolman%20Disease%22)&rank=6 (accessed on 3 January 2023).
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From Production to Applications. Sep. Purif. Rev. 2019, 49, 143–158. [Google Scholar] [CrossRef]
- Akshita, M.; Urgyn, B.; Reena, G. Fungal lipases: A review. J. Biotech Res. 2017, 8, 58–77. [Google Scholar]
- Evaluating the Efficacy of RELiZORB in Managing Exocrine Pancreatic Insufficiency in Tube-Fed Pancreatitis Patients. ClinicalTrials.gov ID NCT06691893. 13.04.2025. Available online: https://www.clinicaltrials.gov/study/NCT06691893?term=AREA%5BBasicSearch%5D(lipase)&rank=9 (accessed on 3 January 2023).
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotech. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef]
- Screening for Lysosomal Acid Lipase Deficiency. ClinicalTrials.gov ID NCT02926872. 16.06.2017. Available online: https://trial.medpath.com/clinical-trial/082237e6fdf79345/nct02926872-screening-lysosomal-acid-lipase-deficiency (accessed on 3 January 2023).
- Lipoprotein Lipase Expression in Chronic Lymphocytic Leukemia. ClinicalTrials.gov ID NCT01460238. 28.03.2019. Available online: https://www.clinicaltrials.gov/study/NCT01460238 (accessed on 3 January 2023).
- Cholinesterase, Amylase, Lipase and Neutrophil-to-Lymphocyte Ratio in Acute Pesticide Poisoning Cases. ClinicalTrials.gov ID NCT05310188. 4.04.2022. Available online: https://aging.networkofcare.org/sanmateo/CommunityResources/ClinicalTrials/Detail/NCT05310188?keyword=%22Amylase%22 (accessed on 3 January 2023).
- Baratta, F.; Pastori, D.; Ferro, D.; Carluccio, G.; Tozzi, G.; Angelico, F.; Violi, F.; Del Ben, M. Reduced lysosomal acid lipase activity: A new marker of liver disease severity across the clinical continuum of non-alcoholic fatty liver disease? World J. Gastroenterol. 2019, 25, 4172–4180. [Google Scholar] [CrossRef]
- Ismail, O.Z.; Bhayana, V. Lipase or amylase for the diagnosis of acute pancreatitis? Clin. Biochem. 2017, 50, 1275–1280. [Google Scholar] [CrossRef]
- Shi, J.; Deng, Q.; Wan, C.; Zheng, M.; Huang, F.; Tang, B. Fluorometric probing of the lipase level as acute pancreatitis biomarkers based on interfacially controlled aggregation-induced emission (AIE). Chem. Sci. 2017, 8, 6188–6195. [Google Scholar] [CrossRef]
- Yun, S. Analysis of Serum Lipase Level Variations in Patients with Pancreatitis. Mod. Gen. Prac. 2023, 1, 8–12. Available online: http://fspress.net/static/upload/file/20240103/1704287230360584.pdf (accessed on 3 January 2023).
- Geldenhuys, W.J.; Lin, L.; Darvesh, A.S.; Sadana, P. Emerging strategies of targeting lipoprotein lipase for metabolic and cardiovascular diseases. Drug Discov. Today 2017, 22, 352–365. [Google Scholar] [CrossRef]
- Hameed, A.M.; Vincent, W.T.; Lam, V.W.T.; Pleass, H.C. Significant elevations of serum lipase not caused by pancreatitis: A systematic review. HPB 2015, 17, 99–112. [Google Scholar] [CrossRef]
- Işık, G.Ç.; Çinpolat, R.; Çevik, Y. Retrospective Analysis of Acute Pancreatitis Cases: Diagnostic Accuracy of Amylase or Lipase Alone. J. Turk. Soc. Rheumatol. 2021, 20, 35–38. [Google Scholar] [CrossRef]
- Anyanwu, G.O.; Kolb, A.F.; Bermano, G. Chapter 9—Antiobesity functional leads and targets for drug development. In Phytochemicals as Lead Compounds for New Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2020; pp. 143–160. [Google Scholar] [CrossRef]
- Mahfoudhi, A.; Benmabrouk, S.; Fendri, A.; Sayari, A. Fungal lipases as biocatalysts: A promising platform in several industrial biotechnology applications. Biotech. Bioeng. 2022, 119, 3370–3392. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Khan, S.A.; Hamayun, M.; Lee, I.-J. The Recent Advances in the Utility of Microbial Lipases: A Review. Microorg. 2023, 11, 510. [Google Scholar] [CrossRef] [PubMed]
- Phulpoto, I.A.; Yu, Z.; Hu, B.; Wang, Y.; Ndayisenga, F.; Li, J.; Liang, H.; Qazi, M.A.; Qazi, M.A. Production and characterization of surfactin-like biosurfactant produced by novel strain Bacillus nealsonii S2MT and it’s potential for oil contaminated soil remediation. Microb. Cell Factories 2020, 19, 145. [Google Scholar] [CrossRef]
- Skoczinski, P.; Volkenborn, K.; Fulton, A.; Bhadauriya, A.; Nutschel, C.; Gohlke, H.; Knapp, A.; Jaeger, K.-E.; Jaeger, K.-E. Contribution of single amino acid and codon substitutions to the production and secretion of a lipase by Bacillus subtilis. Microb. Cell Factories 2017, 16, 160. [Google Scholar] [CrossRef]
- Novy, V.; Carneiro, L.V.; Shin, J.H.; Larsbrink, J.; Olsson, L. Phylogenetic analysis and in-depth characterization of functionally and structurally diverse CE5 cutinases. J. Biol. Chem. 2021, 297, 101302. [Google Scholar] [CrossRef]
- Trail, F.; Köller, W. Diversity of cutinases from plant pathogenic fungi: Purification and characterization of two cutinases from Alternaria brassicicola. Physiol. Mol. Plant Pathol. 1993, 42, 205–220. [Google Scholar] [CrossRef]
- Leger, R.J.S.; Joshi, L.; Roberts, D.W. Adaptation of proteases and carbohydrates of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 1997, 143, 1983–1992. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Chen, J.; Wu, J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013, 31, 1754–1767. [Google Scholar] [CrossRef]
- Dutta, K.; Sen, S.; Veeranki, V.D. Production, characterization and applications of microbial cutinases. Process Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Villafana, R.T.; Rampersad, S.N. Diversity, structure, and synteny of the cutinase gene of Colletotrichum species. Ecol. Evol. 2020, 10, 1425–1443. [Google Scholar] [CrossRef] [PubMed]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Duan, X.; Jiang, Z.; Liu, Y.; Yan, Q.; Xiang, M.; Yang, S. High-level expression of codon-optimized Thielavia terrestris cutinase suitable for ester biosynthesis and biodegradation. Int. J. Biol. Macromol. 2019, 135, 768–775. [Google Scholar] [CrossRef]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. A continuous membrane bioreactor for ester synthesis in organic media: II. Modeling of MBR continuous operation. Biotechnol. Bioeng. 2001, 72, 136–143. [Google Scholar] [CrossRef]
- Rueda Rueda, H.A.; Jimenez-Junca, C.A.; Prieto Correa, R.E. Cutinases obtained from filamentous fungi: Comparison of screening methods. DYNA 2020, 87, 183–190. [Google Scholar] [CrossRef]
- Castro-Ochoa, D.; Peña-Montes, C.; González-Canto, A.; Alva-Gasca, A.; Esquivel-Bautista, R.; Navarro-Ocaña, A.; Farrés, A. ANCUT2, an extracellular cutinase from Aspergillus nidulans induced by olive oil. Appl. Biochem. Biotechnol. 2012, 166, 1275–1290. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Franco, C.F.; Baptista, R.P.; Cabral, J.M.S.; Coelho, A.V.; Rodrigues, C.J.; Melo, E.P. Purification and identification of cutinases from Colletotrichum kahawae and Colletotrichum gloeosporioides. Appl. Microbiol. Biotechnol. 2007, 73, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.S.; Kolattukudy, P.E. Structural studies on cutinase, a glycoprotein containing novel amino acids and glucuronic acid amide at the N terminus. Eur. J. Biochem. 1980, 106, 341–351. [Google Scholar] [CrossRef]
- Weisenborn, P.C.M.; Meder, H.; Egmond, M.R.; Visser, T.J.W.G.; van Hoek, A. Photophysics of the single tryptophan residue in Fusarium solani cutinase: Evidence for the occurrence of conformational substates with unusual fluorescence behaviour. Biophys. Chem. 1996, 58, 281–288. [Google Scholar] [CrossRef]
- Chen, S.; Tong, X.; Woodard, R.W.; Du, G.; Wu, J.; Chen, J. Identification and Characterization of Bacterial Cutinase. J. Biol. Chem. 2008, 283, 25854–25862. [Google Scholar] [CrossRef]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase: From molecular level to bioprocess development. Biotechnol. Bioeng. 1999, 66, 17–34. [Google Scholar] [CrossRef]
- Chen, S.; Su, L.; Billig, S.; Zimmermann, W.; Chen, J.; Wu, J. Biochemical characterization of the cutinases from Thermobifida fusca. J. Mol. Catal. B Enzym. 2010, 63, 121–127. [Google Scholar] [CrossRef]
- Baker, P.J.; Poultney, C.; Liu, Z.; Gross, R.; Montclare, J.K. Identification and comparison of cutinases for synthetic polyester degradation. Appl Micro. Biotech. 2012, 93, 229–240. [Google Scholar] [CrossRef]
- Abokitse, K.; Grosse, S.; Leisch, H.; Corbeil, C.R.; Perrin-Sarazin, F.; Lau, P.C.K. A Novel Actinobacterial Cutinase Containing a Noncatalytic Polymer-Binding Domain. Appl. Environ. Microbiol. 2022, 88, e0152221. [Google Scholar] [CrossRef]
- Sui, B.; Wang, T.; Fang, J.; Hou, Z.; Shu, T.; Lu, Z.; Liu, F.; Zhu, Y. Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes. Front. Microbiol. 2023, 14, 1265139. [Google Scholar] [CrossRef]
- Ferrario, V.; Pellis, A.; Cespugli, M.; Guebitz, G.M.; Gardossi, L. Nature Inspired Solutions for Polymers: Will Cutinase Enzymes Make Polyesters and Polyamides Greener? Catalysts 2016, 6, 205. [Google Scholar] [CrossRef]
- Takahashi, K.; Shimada, T.; Kondo, M.; Tamai, A.; Mori, M.; Nishimura, M.; Hara-Nishimura, I. Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Phys. 2010, 51, 123–131. [Google Scholar] [CrossRef]
- Bååth, J.A.; Novy, V.; Carneiro, L.V.; Guebitz, G.M.; Olsson, L.; Westh, P.; Ribitsch, D. Structure-function analysis of two closely related cutinases from Thermobifida cellulosilytica. Biotec. Bioeng. 2022, 119, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Arya, G.C.; Cohen, H. The Multifaceted Roles of Fungal Cutinases during Infection. J. Fungi 2022, 8, 199. [Google Scholar] [CrossRef]
- Kolattukudy, P.E. Biopolyester Membranes of Plants: Cutin and Suberin. Science 1980, 208, 990–1000. [Google Scholar] [CrossRef]
- Wolfram, K.; Chenglin, Y.; Frances, T.; Parker, D.M. Role of cutinase in the invasion of plants. Can. J. Bot. 1996, 73 (Suppl. 1), 1109–1118. [Google Scholar] [CrossRef]
- Liu, T.; Hou, J.; Wang, Y.; Jin, Y.; Borth, W.; Zhao, F.; Liu, Z.; Hu, J.; Zuo, Y. Genome-wide identification, classification and expression analysis in fungal–plant interactions of cutinase gene family and functional analysis of a putative ClCUT7 in Curvularia lunata. Mol. Genet. Genom. 2016, 291, 1105–1115. [Google Scholar] [CrossRef]
- Egmond, M.R.; de Vlieg, J. Fusarium solani pisi cutinase. Biochimie 2000, 82, 1015–1021. [Google Scholar] [CrossRef]
- Sebastian, J.; Kolattukudy, P.E. Purification and characterization of cutinase from a fluorescent Pseudomonas putida bacterial strain isolated from phyllosphere. Arch. Biochem. Biophys. 1988, 263, 77–85. [Google Scholar] [CrossRef]
- De Jesus, R.; Alkendi, R. A minireview on the bioremediative potential of microbial enzymes as solution to emerging microplastic pollution. Front. Microbiol. 2023, 13, 1066133. [Google Scholar] [CrossRef]
- Bhandari, G. Mycoremediation: An Eco-friendly Approach for Degradation of Pesticides. In Mycoremediation and Environmental Sustainability; Prasad, R., Ed.; Fungal Biology; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.A.; Khalifa, H.O.; Yoon, H.J.; Ki, M.-R.; Pack, S.P. Microbial Immobilized Enzyme Biocatalysts for Multipollutant Mitigation: Harnessing Nature’s Toolkit for Environmental Sustainability. Int. J. Mol. Sci. 2024, 25, 8616. [Google Scholar] [CrossRef] [PubMed]
- Satti, S.M.; Shah, A.A. Polyester-based biodegradable plastics: An approach towards sustainable development. Lett. Appl. Microbiol. 2020, 70, 413–430. [Google Scholar] [CrossRef]
- Nyyssölä, A. Which properties of cutinases are important for applications? Appl. Microbiol. Biotechnol. 2015, 99, 4931–4942. [Google Scholar] [CrossRef]
- Ahn, J.-Y.; Kim, Y.-H.; Min, J.; Lee, J. Accelerated degradation of dipentyl phthalate by Fusarium oxysporum f. sp. pisi cutinase and toxicity evaluation of its degradation products using bioluminescent bacteria. Curr. Microbiol. 2006, 52, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Lee, J.; Ahn, J.-Y.; Gu, M.B.; Moon, S.-H. Enhanced degradation of an endocrine-disrupting chemical, butyl benzyl phthalate, by Fusarium oxysporum f. sp. pisi cutinase. Appl. Environ. Microbiol. 2002, 68, 4684–4688. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, J.; Moon, S.H. Degradation of an endocrine disrupting chemical, DEHP [di-(2-ethylhexyl)-phthalate], by Fusarium oxysporum f. sp. pisi cutinase. Appl. Microbiol. Biotechnol. 2003, 63, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Teles, F.R.R.; Cabral, J.M.S.; Santos, J.A.L. Enzymatic degreasing of a solid waste from the leather industry by lipases. Biotechnol. Lett. 2001, 23, 1159–1163. [Google Scholar] [CrossRef]
- Gururaj, P.; Khushbu, S.; Monisha, B.; Selvakumar, N.; Chakravarthy, M.; Gautam, P.; Nandhini Devi, G. Production, purification and application of cutinase in enzymatic scouring of cotton fabric isolated from Acinetobacter baumannii AU10. Prep. Biochem. Biotechnol. 2021, 51, 550–561. [Google Scholar] [CrossRef]
- Degani, O. Synergism between cutinase and pectinase in the hydrolysis of cotton fibers’ cuticle. Catalysts 2021, 11, 84. [Google Scholar] [CrossRef]
- Poulose, A.J.; Boston, M. Enzyme Assisted Degradation of Surface Membranes of Harvested Fruits and Vegetables. U.S. Patent 5037662 A, 23 June 1996. [Google Scholar]
- Rinaldi, S.; Van der Kamp, M.W.; Ranaghan, K.E.; Mulholland, A.J.; Colombo, G. Understanding complex mechanisms of enzyme reactivity: The case of limonene-1,2-epoxide hydrolases. Catalysts 2018, 8, 5698–5707. [Google Scholar] [CrossRef]
- Su, L.; Hong, R.; Kong, D.; Wu, J. Enhanced activity towards polyacrylates and poly(vinyl acetate) by site-directed mutagenesis of Humicola insolens cutinase. Int. J. Biol. Macromol. 2020, 162, 1752–1759. [Google Scholar] [CrossRef]
- Chen, C.-C.; Dai, L.; Ma, L.; Guo, R.-T. Enzymatic degradation of plant biomass and synthetic polymers. Nat. Rev. Chem. 2020, 4, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Carniel, A.; Gomes, A.D.C.; Coelho, M.A.Z.; de Castro, A.M. Process strategies to improve biocatalytic depolymerization of post-consumer PET packages in bioreactors, and investigation on consumables cost reduction. Bioprocess Biosyst. Eng. 2021, 44, 507–516. [Google Scholar] [CrossRef]
- Sankhla, I.S.; Sharma, G.; Tak, A. Fungal degradation of Bioplastics: An overview. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 35–47. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustain. Chem. Eng. 2020, 8, 8894–8908. [Google Scholar] [CrossRef]
- Vogel, K.; Wei, R.; Pfaff, L.; Breite, D.; Al-Fathi, H.; Ortmann, C.; Estrela-Lopis, I.; Venus, T.; Schulze, A.; Harms, H.; et al. Enzymatic degradation of polyethylene terephthalate nanoplastics analyzed in real time by isothermal titration calorimetry. Sci. Total Environ. 2021, 773, 145111. [Google Scholar] [CrossRef]
- Huang, S.J. Polymer Waste Management–Biodegradation, Incineration, and Recycling. J. Macromol. Sci. Part A Pure Appl. Chem. 1995, 32, 593–597. [Google Scholar] [CrossRef]
- Shi, K.; Jing, J.; Song, L.; Su, T.; Wang, Z. Enzymatic hydrolysis of polyester: Degradation of poly(ε-caprolactone) by Candida antarctica lipase and Fusarium solani cutinase. Int. J. Biol. Macromol. 2020, 144, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Moeis, M.R.; Maulana, M.F. Improving plastic degradation by increasing the thermostability of a whole cell biocatalyst with LC-cutinase activity. J. Phys. Conf. Ser. 2021, 1764, 012029. [Google Scholar] [CrossRef]
- Akçaözoğlu, S.; Adıgüzel, A.O.; Akçaözoğlu, K.; Deveci, E.Ü.; Gönen, Ç. Investigation of the bacterial modified waste PET aggregate via Bacillus safensis to enhance the strength properties of mortars. Constr. Build. Mater. 2021, 270, 121828. [Google Scholar] [CrossRef]
- Kawai, F. The current state of research on PET hydrolyzing enzymes available for biorecycling. Catalysts 2021, 11, 206. [Google Scholar] [CrossRef]
- Dimarogona, M.; Nikolaivits, E.; Kanelli, M.; Christakopoulos, P.; Sandgren, M.; Topakas, E. Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential. Biochim. Bioph. Acta 2015, 1850, 2308–2317. [Google Scholar] [CrossRef]
- Singhvi, M.S.; Zinjarde, S.S.; Gokhale, D.V. Polylactic acid: Synthesis and biomedical applications. J. Appl. Microbiol. 2019, 127, 1612–1626. [Google Scholar] [CrossRef]
- Kim, M.Y.; Kim, C.; Moon, J.; Heo, J.; Jung, S.P.; Kim, J.R. Polymer film-based screening and isolation of polylactic acid (PLA)-degrading microorganisms. J. Microbiol. Biotechnol. 2017, 27, 342–349. [Google Scholar] [CrossRef]
- Kitadokoro, K.; Kakara, M.; Matsui, S.; Osokoshi, R.; Thumarat, U.; Kawai, F.; Kamitani, S. Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase. FEBS J. 2019, 286, 2087–2098. [Google Scholar] [CrossRef]
- Fortuna, S.; Cespugli, M.; Todea, A.; Pellis, A.; Gardossi, L. Criteria for Engineering Cutinases: Bioinformatics Analysis of Catalophores. Catalysts 2021, 11, 784. [Google Scholar] [CrossRef]
- Thapa, S.; Li, H.; Ohair, J.; Bhatti, S.; Chen, F.-C.; Al Nasr, K.; Johnson, T.; Zhou, S. Biochemical Characteristics of Microbial Enzymes and Their Significance from Industrial Perspectives. Mol. Biotech. 2019, 61, 579–601. [Google Scholar] [CrossRef]
- Ronkvist, A.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-Catalyzed Hydrolysis of Poly(ethylene terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Ping, L.-F.; Chen, X.; Yuan, X.; Zhang, M.; Chai, Y.; Shan, S. Application and comparison in biosynthesis and biodegradation by Fusarium solani and Aspergillus fumigatus cutinases. Inter. J. Bio. Macromol. 2017, 104 Pt A, 1238–1245. [Google Scholar] [CrossRef]
- Puchalski, M.; Szparaga, G.; Biela, T.; Gutowska, A.; Sztajnowski, S.; Krucińska, I. Molecular and supramolecular changes in polybutylene succinate (PBS) and polybutylene succinate adipate (PBSA) copolymer during degradation in various environmental conditions. Polymers 2018, 10, 251. [Google Scholar] [CrossRef]
- Tan, Y.; Henehan, G.T.; Kinsella, G.K.; Ryan, B.J. An extracellular lipase from Amycolatopsis mediterannei is a cutinase with plastic degrading activity. Comp. Struct. Biotechnol. J. 2021, 19, 869–879. [Google Scholar] [CrossRef]
- Aloulou, A.; Rahier, R.; Arhab, Y.; Noiriel, A.; Abousalham, A. Phospholipases: An Overview. In Lipases and Phospholipases; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1835. [Google Scholar] [CrossRef]
- Murakami, M.; Nakatani, Y.; Atsumi, G.; Inoue, K.; Kudo, I. Regulatory Functions of Phospholipase A2. Crit. Rev. Immun. 2017, 37, 127–195. [Google Scholar] [CrossRef]
- O’Donnell, V.B.; Rossjohn, J.; Wakelam, M.J.O. Phospholipid signaling in innate immune cells. J. Clin. Investig. 2018, 128, 2670–2679. [Google Scholar] [CrossRef]
- Murakami, M. Lipoquality control by phospholipase A2 enzymes. Proc. Jpn. Acad. Ser. B 2017, 93, 677–702. [Google Scholar] [CrossRef]
- Bill, C.A.; Vines, C.M. Phospholipase C. In Calcium Signaling; Islam, M., Ed.; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2020; Volume 1131. [Google Scholar] [CrossRef]
- Salucci, S.; Aramini, B.; Bartoletti-Stella, A.; Versari, I.; Martinelli, G.; Blalock, W.; Stella, F.; Faenza, I. Phospholipase Family Enzymes in Lung Cancer: Looking for Novel Therapeutic Approaches. Cancers 2023, 15, 3245. [Google Scholar] [CrossRef] [PubMed]
- Balboa, M.A.; Balsinde, J. Phospholipases: From Structure to Biological Function. Biomolecules 2021, 11, 428. [Google Scholar] [CrossRef] [PubMed]
- De Barros, D.P.C.; Fonseca, L.P.; Fernandes, P.; Cabral, J.M.S.; Mojovic, L. Biosynthesis of ethyl caproate and other short ethyl esters catalyzed by cutinase in organic solvent. J. Mol. Cat. B Enzym. 2009, 60, 178–185. [Google Scholar] [CrossRef]
- Dutta, K.; Dasu, V.V. Synthesis of short chain alkyl esters using cutinase from Burkholderia cepacia NRRL B2320. J. Mol. Cat. B Enzym. 2011, 72, 150–156. [Google Scholar] [CrossRef]
- Nelson, R.K.; Frohman, M.A. Thematic Review Series: Phospholipases: Central Role in Lipid Signaling and Disease. J. Lip. Res. 2015, 56, 2229–2237. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, L.; Gao, J.; Li, Z.; Jäckering, A.; Weber, G.; Mican, J.; Chen, Y.; Dong, W.; Han, X.; Feiler, C.G.; et al. Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Cat. 2022, 12, 9790–9800. [Google Scholar] [CrossRef] [PubMed]
- Alekseeva, A.S.; Boldyrev, I.A. Phospholipase A2. Methods for Activity Monitoring. Biochem. Moscow Suppl. Ser. A 2020, 14, 267–278. [Google Scholar] [CrossRef]
- El Alaoui, M.; Soulère, L.; Noiriel, A.; Popowycz, F.; Khatib, A.; Queneau, Y.; Abousalham, A. A continuous spectrophotometric assay that distinguishes between phospholipase A1 and A2 activities. J. Lipid Res. 2016, 57, 1589–1597. [Google Scholar] [CrossRef]
- Garcia, A.; Deplazes, E.; Aili, S.; Padula, M.P.; Touchard, A.; Murphy, C.; Lankage, U.M.; Nicholson, G.M.; Cornell, B.; Cranfield, C.G. Label-Free, Real-Time Phospholipase—A Isoform Assay. ACS Biomater. Sci. Eng. 2020, 6, 4714–4721. [Google Scholar] [CrossRef]
- Zhang, Y.; Ai, J.; Dong, Y.; Zhang, S.; Gao, O.; Qi, H.; Zhang, C.; Cheng, Z. Combining 3D graphene-like screen-printed carbon electrode with methylene blue-loaded liposomal nanoprobes for phospholipase A2 detection. Biosens. Bioelectron. 2019, 126, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Rahier, R.; Noiriel, A.; Abousalham, A. Development of a Direct and Continuous Phospholipase D Assay Based on the Chelation-Enhanced Fluorescence Property of 8-Hydroxyquinoline. Anal. Chem. 2016, 88, 666–674. [Google Scholar] [CrossRef]
- Chapman, R.; Lin, Y.; Burnapp, M.; Bentham, A.; Hillier, D.; Zabron, A.; Khan, S.; Tyreman, M.; Stevens, M.M. Multivalent Nanoparticle Networks Enable Point-of-Care Detection of Human Phospholipase-A2 in Serum. ACS Nano 2015, 9, 2565–2573. [Google Scholar] [CrossRef]
- Bohr, S.S.-R.; Thorlaksen, C.; Kühnel, R.M.; Günther-Pomorski, T.; Hatzakis, N.S. Label-Free Fluorescence Quantification of Hydrolytic Enzyme Activity on Native Substrates Reveals How Lipase Function Depends on Membrane Curvature. Langmuir 2020, 36, 6473–6481. [Google Scholar] [CrossRef]
- Su, A.; Kiokekli, S.; Naviwala, M.; Shirke, A.N.; Pavlidis, I.V.; Gross, R.A. Cutinases as stereoselective catalysts: Specific activity and enantioselectivity of cutinases and lipases for menthol and its analogs. Enzyme Microb. Technol. 2020, 133, 109467. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef]
- Saavedra, D.E.M.; Baltar, F. Multifunctionality of alkaline phosphatase in ecology and biotechnology. Curr. Opin. Biotechnol. 2025, 91, 103229. [Google Scholar] [CrossRef]
- Cerminati, S.; Paoletti, L.; Aguirre, A.; Peirú, S.; Menzella, H.G.; Castelli, M.E. Industrial uses of phospholipases: Current state and future applications. Appl. Microbiol. Biotechnol. 2019, 103, 2571–2582. [Google Scholar] [CrossRef]
- Smith, M.R.; Khera, E.; Wen, F. Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res. 2015, 54, 4021–4032. [Google Scholar] [CrossRef]
- Tsai, S.-L.; DaSilva, N.A.; Chen, W. Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth. Biol. 2013, 2, 14–21. [Google Scholar] [CrossRef]
- Casado, V.; Martín, D.; Torres, C.; Reglero, G. Phospholipases in food industry: A review. Methods Mol. Biol. 2012, 861, 495–523. [Google Scholar] [CrossRef]
- Ma, Y.; Yao, M.; Li, B.; Ding, M.; He, B.; Chen, S.; Zhou, X.; Yuan, Y.; Yuan, Y. Enhanced Poly(ethylene terephthalate) hydrolase Activity by Protein Engineering. Engineering 2018, 4, 888–893. [Google Scholar] [CrossRef]
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Clarke, D.J.; Dobson, A.D.W. Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Front. Microbiol. 2020, 11, 571265. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019, 103, 4253–4268. [Google Scholar] [CrossRef]
- Han, X.; Liu, W.; Huang, J.-W.; Ma, J.; Zheng, Y.; Ko, T.-P.; Xu, L.; Cheng, Y.-S.; Chen, C.-C.; Guo, R.-T.; et al. Structural insight into catalytic mechanism of PET hydrolase. Nat. Commun. 2017, 8, 2106. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; He, L.; Wang, L.; Li, T.; Li, C.; Liu, H.; Luo, Y.; Bao, R.; Bao, R. Cover Feature: Protein crystallography and Site-Direct Mutagenesis Analysis of the Poly(ethylene terephthalate) hydrolase PETase from Ideonella sakaiensis (ChemBioChem 14/2018). ChemBioChem 2018, 19, 1464. [Google Scholar] [CrossRef]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current status and application aspects. ACS Cat. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Herrero Acero, E.; Ribitsch, D.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Steinkellner, G.; Gruber, K.; Schwab, H.; Guebitz, G.M.; Schwab, H.; et al. Surface engineering of a cutinase from Thermobifida cellulosilytica for improved polyester hydrolysis. Biotechnol. Bioeng. 2013, 110, 2581–2590. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Da, S.; Silva, N.; Matamá, T.; Araújo, R.; Martins, M.; Chen, S.; Chen, J.; Wu, J.; Casal, M.; et al. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates. Biotechnol. J. 2011, 6, 1230–1239. [Google Scholar] [CrossRef]
- Endo, Y. Development of a cell-free protein synthesis system for practical use. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021, 97, 261–276. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational Protein Engineering of Thermo-Stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X.; et al. Computational Redesign of a PETase for Plastic Biodegradation under Ambient Condition by the GRAPE Strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.R.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Sevilla, M.E.; Garcia, M.D.; Perez-Castillo, Y.; Armijos-Jaramillo, V.; Casado, S.; Vizuete, K.; Debut, A.; Cerda-Mejía, L. Degradation of PET Bottles by an Engineered Ideonella sakaiensis PETase. Polymers 2023, 15, 1779. [Google Scholar] [CrossRef]
- Barclay, A.; Acharya, K.R. Engineering Plastic Eating Enzymes Using Structural Biology. Biomolecules 2023, 13, 1407. [Google Scholar] [CrossRef]
- Brott, S.; Pfaff, L.; Schuricht, J.; Schwarz, J.; Böttcher, D.; Badenhorst, C.P.S.; Wei, R.; Bornscheuer, U.T. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Eng. Life Sci. 2022, 22, 192–203. [Google Scholar] [CrossRef]
- Kawai, F. Emerging Strategies in Polyethylene Terephthalate Hydrolase Research for Biorecycling. ChemSusChem 2021, 14, 4115. [Google Scholar] [CrossRef] [PubMed]
- Ermis, H. A mini-review on the role of PETase in polyethylene terephthalate degradation. Rev. Environ. Sci. Biotechnol. 2025, 24, 545–555. [Google Scholar] [CrossRef]
- Almeida, E.L.; Carrillo Rincón Andrés, F.C.R.; Jackson, S.A.; Dobson, A.D.W. In silico Screening and Heterologous Expression of a Polyethylene Terephthalate Hydrolase (PETase)-like Enzyme (SM14est) with Polycaprolactone (PCL)-Degrading Activity, from the Marine Sponge-Derived Strain Streptomyces sp. SM14. Front. Microb. 2019, 10, 2187. [Google Scholar] [CrossRef] [PubMed]
- Khairul Anuar, N.F.S.; Huyop, F.; Ur-Rehman, G.; Abdullah, F.; Normi, Y.M.; Sabullah, M.K.; Abdul Wahab, R. An Overview into Polyethylene Terephthalate (PET) Hydrolases and Efforts in Tailoring Enzymes for Improved Plastic Degradation. Int. J. Mol. Sci. 2022, 23, 12644. [Google Scholar] [CrossRef]
- Jin, J.; Jia, Z. Characterization of Potential Plastic-Degradation Enzymes from Marine Bacteria. ACS Omega 2024, 9, 32185–32192. [Google Scholar] [CrossRef]
- Choi, J.-M. Plastic-Degrading Enzymes as Sustainable Solutions for Plastic Waste. Preprints 2025, 2025040667. Available online: https://sciety.org/articles/activity/10.20944/preprints202504.0667.v1 (accessed on 3 January 2023).
- Chen, K.; Hu, Y.; Dong, X.; Sun, Y. Molecular Insights into the Enhanced Performance of EKylated PETase Toward PET Degradation. ACS Catal. 2021, 11, 7358–7370. [Google Scholar] [CrossRef]
- Choi, J.; Kim, H.; Ahn, Y.-R.; Kim, M.; Yu, S.; Kim, N.; Lim, S.Y.; Park, J.-A.; Ha, S.-J.; Lim, K.S.; et al. Recent advances in microbial and enzymatic engineering for the biodegradation of micro- and nanoplastics. RSC Adv. 2024, 14, 9943–9966. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Yan, W.; Cao, Z.; Ding, M.; Yuan, Y. Current Advances in the Biodegradation and Bioconversion of Polyethylene Terephthalate. Microorganisms 2022, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Sun, S. Recent advances in screening and identification of PET-degrading enzymes. Environ. Rev. 2024, 32, 294–314. [Google Scholar] [CrossRef]
- Ahmaditabatabaei, S.; Kyazze, G.; Iqbal, H.M.N.; Keshavarz, T. Fungal Enzymes as Catalytic Tools for Polyethylene Terephthalate (PET) Degradation. J. Fungi 2021, 7, 931. [Google Scholar] [CrossRef]
- Jayasekara, S.K.; Joni, H.D.; Jayantha, B.; Dissanayake, L.; Mandrell, C.; Sinharage, M.M.; Molitor, R.; Jayasekara, T.; Sivakumar, P.; Jayakody, L.N. Trends in in-silico guided engineering of efficient polyethylene terephthalate (PET) hydrolyzing enzymes to enable bio-recycling and upcycling of PET. Comput. Struct. Biotechnol. J. 2023, 21, 3513–3521. [Google Scholar] [CrossRef]
- Li, S. Application of PETase in Plastic Biodegradation and Its Synthesis. E3S Web Conf. 2024, 553, 03015. [Google Scholar] [CrossRef]
- Martín-González, D.; de la Fuente Tagarro, C.; De Lucas, A.; Bordel, S.; Santos-Beneit, F. Genetic Modifications in Bacteria for the Degradation of Synthetic Polymers: A Review. Int. J. Mol. Sci. 2024, 25, 5536. [Google Scholar] [CrossRef]
- Ogunlusi, T.S.; Ikoyo, S.S.; Dadashipour, M.; Gao, H. Engineering Is PETase and Its Homologues: Advances in Enzyme Discovery and Host Optimisation. Int. J. Mol. Sci. 2025, 26, 6797. [Google Scholar] [CrossRef]


| Type of Esterase | Microorganism | Substarte | Optimal pH | Optimal Temperature (°C) | Molecular Weight (kDa) | Km (µM) | Vmax | Application |
|---|---|---|---|---|---|---|---|---|
| (µM/min/mg) | ||||||||
| Serine esterase | Aspergillus westerdijkiae | Water-soluble short-chain fatty acids | 8 | 40 | 32 | 638.11 | 5.47 | As a potential biotechnological catalyst |
| Hydrolysis of hemicellulose and lignin | ||||||||
| Glucuronoyl esterase | Aspergillus fumigatus | Hemicellulose and lignin | 5 | 40 a 50 | Nd | 15.8 y | Nd | |
| (Favorable) | 16.4 | Lignocellulose hydrolysis | ||||||
| Glucuronoyl esterase | Neurospora crassa | Lignocellulose | 7 | 40 a 50 | 32 | 15 | 1.12 | |
| (Favorable) | ||||||||
| 1.0 | Synthesis of flavor esters | |||||||
| Recombinant esterase (RmEstA) | Rhizomucor miehei | pNP with acyl lengths from C2 to C16 | 6.5 | 45 | 34 | 0.17 | Nd | |
| 0.12 | ||||||||
| 0.82 | ||||||||
| 0.28 y | ||||||||
| 0.3 | ||||||||
| Feruloyl esterase (Est1) | Pleurotus sapidus | Feruloylated saccharides | 6 | 50 | 55 | 1.95 | 1.77 | Ecological or technical applications |
| EstS1 | Sulfobacillus acidophilus | Phthalates | 8 | 70 | 36 | 0.18 | 2440 | Degradation of phthalates |
| Lp-1002 esterase | Lactobacillus plantarum (WCFS1) | Phenyl acetate | 5-7 | 40 | Nd | Nd | Nd | Winemaking |
| EstB28 | Oenococcus oeni | Nitrophenyl-linked substrates | 5 | 40 | 34.5 | Nd | Nd | Winemaking |
| CL96 esterase | Lactobacillus casei | ρ-Nitrophenyl derivatives of fatty acids (C2 and C4) | 7 | 30 | Nd | Nd | Nd | Dairy industry |
| Cholesterol esterase | Pseudomonas aeruginosa | Fatty acid cholesteryl | 7 | 53 | 58 | Nd | Nd | Optical industry |
| Cinnamoyl esterase | Lactobacillus helveticus (KCCM 11223) | Methyl ferulate, methyl sinapinate, methyl ρ-coumarate and methyl caffeate | 7 | 65 | 27.4 | 0.153 | 559.6 | Hydrolysis of chlorogenic acid |
| LipM | Metagenomics of agricultural soil expressed in Escherichia coli | ρ-nitrophenyl short-chain fatty acids | 7.5 | 37 | 48 | Nd | Nd | Transesterification of polluting compounds, production of biodiesel or food supplement for monogastric animals |
| S. cerevisiae esterase | Saccharomyces cerevisiae | Formaldehyde | 7 | 50 | 40 | 0.29 | 12 | Formaldehyde detoxification |
| Microorganism | Temperature (°C) | pH | kcat (s−1) | KM (mM) | Kcat/KM (M−1s−1) | References |
|---|---|---|---|---|---|---|
| Wild-type IsPETase | 30 | 7.5 | ~0.1 | 1.5–4.0 | 20–80 | [161,176] |
| S238F/ W159H | 30 | 7.5 | 0.2–0.5 | 0.4–1.0 | 500–1200 | [176] |
| Thermo PETase | 40 | 8.0 | 0.8–1.2 | 0.3–0.8 | 1500–2500 | [177] |
| Dura PETase | 40 | 8.0 | 1.0–1.5 | 0.3–0.5 | 2000–3500 | [178] |
| FAST- PETase | 50 | 8.0 | 2.0–3.0 | 0.3–0.5 | 6000–7000 | [178] |
| Hot PETase | 60 | 8.5 | 2.5–4.0 | 0.3–0.6 | 6000–8000 | [179] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mussakhmetov, A.; Silayev, D. Esterases: Mechanisms of Action, Biological Functions, and Application Prospects. Appl. Microbiol. 2025, 5, 139. https://doi.org/10.3390/applmicrobiol5040139
Mussakhmetov A, Silayev D. Esterases: Mechanisms of Action, Biological Functions, and Application Prospects. Applied Microbiology. 2025; 5(4):139. https://doi.org/10.3390/applmicrobiol5040139
Chicago/Turabian StyleMussakhmetov, Arman, and Dmitriy Silayev. 2025. "Esterases: Mechanisms of Action, Biological Functions, and Application Prospects" Applied Microbiology 5, no. 4: 139. https://doi.org/10.3390/applmicrobiol5040139
APA StyleMussakhmetov, A., & Silayev, D. (2025). Esterases: Mechanisms of Action, Biological Functions, and Application Prospects. Applied Microbiology, 5(4), 139. https://doi.org/10.3390/applmicrobiol5040139








