Author Contributions
The work has been contributed by the authors as follows: Conceptualization, M.A., A.B., H.M. and H.K.T.; methodology, H.K.T.; software, H.K.T.; validation, H.K.T.; formal analysis, E.F.S., Z.G., G.S., M.A., A.B., H.M. and H.K.T.; investigation, E.F.S., Z.G., G.S., M.A., A.B., H.M. and H.K.T.; resources, M.A., A.B. and H.M.; data curation, H.K.T.; writing—original draft preparation, E.F.S., Z.G., M.A. and H.K.T.; writing—review and editing, E.F.S., Z.G. and H.K.T.; visualization, Z.G. and H.K.T.; supervision, G.S., Z.G. and H.K.T.; project administration, H.K.T.; funding acquisition, H.K.T. All authors have read and agreed to the published version of the manuscript.
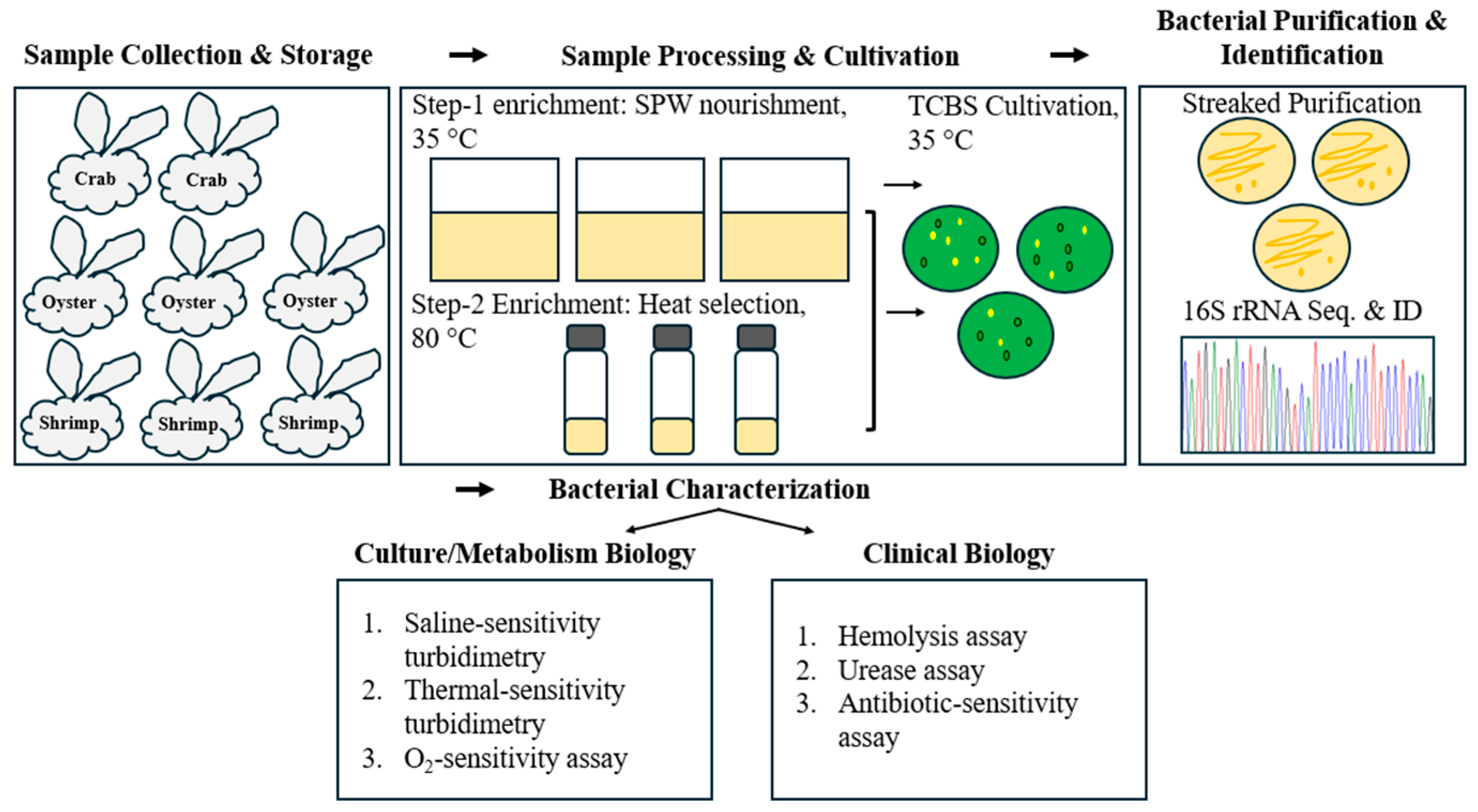
Figure 1.
Experimental scheme for the exploration of V. lutrae.
Figure 1.
Experimental scheme for the exploration of V. lutrae.
Figure 2.
Yellow regular (panel A) and heat-resistant (panel B) V. lutrae colony morphologies on TCBS agar. Enriched seafood solutions containing presumptive V. lutrae were inoculated on TCBS agar and incubated at 37 °C until colorful colonies (i.e., green and yellow colors) emerged. Yellow colonies were purified, and V. lutrae was confirmed using 16S rRNA bacterial identification.
Figure 2.
Yellow regular (panel A) and heat-resistant (panel B) V. lutrae colony morphologies on TCBS agar. Enriched seafood solutions containing presumptive V. lutrae were inoculated on TCBS agar and incubated at 37 °C until colorful colonies (i.e., green and yellow colors) emerged. Yellow colonies were purified, and V. lutrae was confirmed using 16S rRNA bacterial identification.
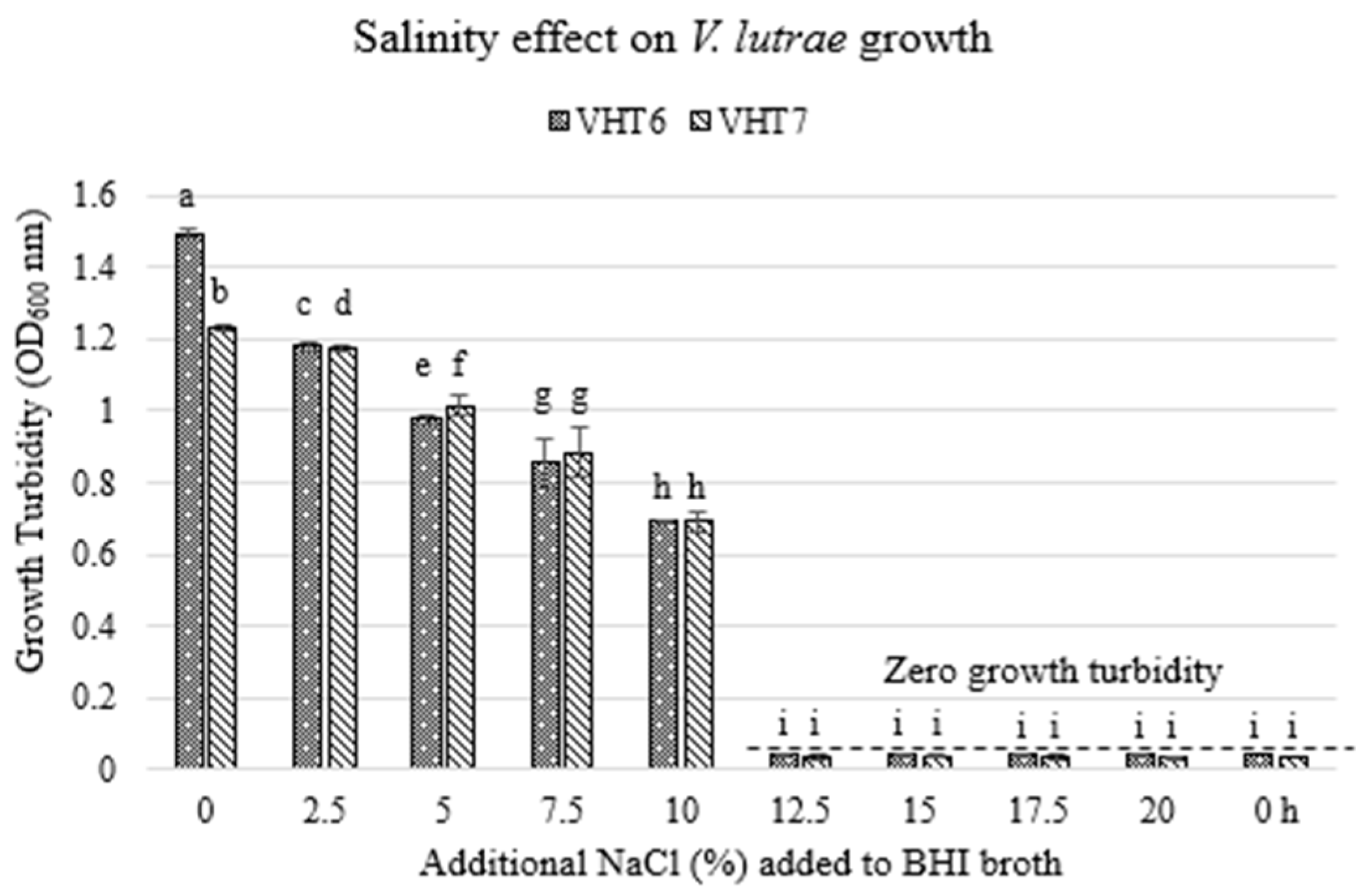
Figure 3.
Growth turbidity of select V. lutrae (i.e., VHT6 and VHT7) in NaCl-supplemented BHI broth. The growth turbidity of bacterial broth cultures at post-incubation (35 °C, 0 h, 24 h, 48 h) was recorded at OD600. Zero growth turbidity was dictated with reference to inoculated BHI broth at zero incubation time (i.e., 0 h). Data bars (i.e., 48 h data only) represent the mean of four replications. Means that share the same lowercase letters are not significantly different; means with different letters are significantly different (p < 0.05). The error bars indicate standard deviation from the mean. Dashed line, zero growth turbidity.
Figure 3.
Growth turbidity of select V. lutrae (i.e., VHT6 and VHT7) in NaCl-supplemented BHI broth. The growth turbidity of bacterial broth cultures at post-incubation (35 °C, 0 h, 24 h, 48 h) was recorded at OD600. Zero growth turbidity was dictated with reference to inoculated BHI broth at zero incubation time (i.e., 0 h). Data bars (i.e., 48 h data only) represent the mean of four replications. Means that share the same lowercase letters are not significantly different; means with different letters are significantly different (p < 0.05). The error bars indicate standard deviation from the mean. Dashed line, zero growth turbidity.
Figure 4.
Viable count (CFU) of NaCl-challenged V. lutrae (i.e., VHT6 and VHT7). Incubated NaCl-challenged cells (i.e., 35 °C, 48 h incubation; 10−3 dilutions) were assessed for viable plate count (i.e., colony formation). The colony detection limit represents viable counts (i.e., CFU) below bacterial countable number (i.e., <30 colonies, too few to count). Data bars represent the mean of four replications. Means that share the same lowercase letters are not significantly different; means with different letters are significantly different (p < 0.05). The error bars indicate standard deviation from the mean. Dashed line, colony detection limit.
Figure 4.
Viable count (CFU) of NaCl-challenged V. lutrae (i.e., VHT6 and VHT7). Incubated NaCl-challenged cells (i.e., 35 °C, 48 h incubation; 10−3 dilutions) were assessed for viable plate count (i.e., colony formation). The colony detection limit represents viable counts (i.e., CFU) below bacterial countable number (i.e., <30 colonies, too few to count). Data bars represent the mean of four replications. Means that share the same lowercase letters are not significantly different; means with different letters are significantly different (p < 0.05). The error bars indicate standard deviation from the mean. Dashed line, colony detection limit.
Figure 5.
Growth turbidity of pasteurized V. lutrae. V. lutrae-inoculated BHI broth tubes (A) were visually evaluated for growth turbidity at post-sequential pasteurization incubation (i.e., 63 °C, 8 h) and culture incubation (i.e., 35 °C, 16 h) relative to non-inoculated BHI broth (B).
Figure 5.
Growth turbidity of pasteurized V. lutrae. V. lutrae-inoculated BHI broth tubes (A) were visually evaluated for growth turbidity at post-sequential pasteurization incubation (i.e., 63 °C, 8 h) and culture incubation (i.e., 35 °C, 16 h) relative to non-inoculated BHI broth (B).
Figure 6.
Viability profile of pasteurized, heat-resistant V. lutrae. Following pasteurization treatment (i.e., 63 °C, 8 h) and culture incubation of inoculated broth, V. lutrae VHT27 (panel A), VHT28 (panel B), and VHT29 (panel C) dilutions were inoculated on agar plates and anaerobically incubated (35 °C) until colonies formed. Using 16S rRNA gene sequencing, bacterial colonies were analyzed and confirmed as V. lutrae, and VHT27, VHT28, and VHT29 were renamed as VHT82, VHT83, and VHT84, respectively.
Figure 6.
Viability profile of pasteurized, heat-resistant V. lutrae. Following pasteurization treatment (i.e., 63 °C, 8 h) and culture incubation of inoculated broth, V. lutrae VHT27 (panel A), VHT28 (panel B), and VHT29 (panel C) dilutions were inoculated on agar plates and anaerobically incubated (35 °C) until colonies formed. Using 16S rRNA gene sequencing, bacterial colonies were analyzed and confirmed as V. lutrae, and VHT27, VHT28, and VHT29 were renamed as VHT82, VHT83, and VHT84, respectively.
Figure 7.
Resuscitation profile of VBNC V. lutrae at post pasteurization (63 °C). Following pasteurization (i.e., 63 °C, 90 min/8 h) treatment and culture incubation of inoculated broth, V. lutrae VHT82 (VHT27 derivative, 63 °C, 90 min) turbid broth (panel A) and VHT44 (63 °C, 8 h) turbid broth (panel B) and viable plating incubated anaerobically (panel D) (35 °C, until colonies formed) were recorded. Non-turbid broth containing VBNC VHT44 and VHT82 cells is represented in (panel C). Upon 16S rRNA gene sequencing of resuscitated VHT44 colonies, it was renamed as VHT85.
Figure 7.
Resuscitation profile of VBNC V. lutrae at post pasteurization (63 °C). Following pasteurization (i.e., 63 °C, 90 min/8 h) treatment and culture incubation of inoculated broth, V. lutrae VHT82 (VHT27 derivative, 63 °C, 90 min) turbid broth (panel A) and VHT44 (63 °C, 8 h) turbid broth (panel B) and viable plating incubated anaerobically (panel D) (35 °C, until colonies formed) were recorded. Non-turbid broth containing VBNC VHT44 and VHT82 cells is represented in (panel C). Upon 16S rRNA gene sequencing of resuscitated VHT44 colonies, it was renamed as VHT85.
Figure 8.
Oxygen-dependent growth of V. lutrae. Layer cultivation (Panel A): VHT82, VHT83, VHT84, VHT85; parafilm-wrapped cultivation (Panel B): VHT83, VHT84, VHT85, VHT28, VHT29; Anaerobic cultivation (Panel C): VHT82. VHT28 (panel A) and VHT29 (panel B) are the native cultures of VHT83 and VHT84, respectively. x, no visible CFUs; v, visible CFUs were detected.
Figure 8.
Oxygen-dependent growth of V. lutrae. Layer cultivation (Panel A): VHT82, VHT83, VHT84, VHT85; parafilm-wrapped cultivation (Panel B): VHT83, VHT84, VHT85, VHT28, VHT29; Anaerobic cultivation (Panel C): VHT82. VHT28 (panel A) and VHT29 (panel B) are the native cultures of VHT83 and VHT84, respectively. x, no visible CFUs; v, visible CFUs were detected.
Figure 9.
Oxygen-dependent growth behavior of V. lutrae in broth media (i.e., 35 °C, 20 h). VHT27 (panel A), VHT28 (panel B), and VHT29 (panel C) are the native cultures of VHT82, VHT83, and VHT84, respectively, and they exhibited non-planktonic growth behavior.
Figure 9.
Oxygen-dependent growth behavior of V. lutrae in broth media (i.e., 35 °C, 20 h). VHT27 (panel A), VHT28 (panel B), and VHT29 (panel C) are the native cultures of VHT82, VHT83, and VHT84, respectively, and they exhibited non-planktonic growth behavior.
Figure 10.
Antibiotic sensitivity profile of select V. lutrae (i.e., VHT84 and VHT85). Each pair of lower/lower and lower/upper case letters represents a non-significant (p > 0.05) and significant (p < 0.05) difference in antibiotic sensitivity, respectively, among V. lutrae strains examined. Data bars (i.e., 48 h data only) represent the mean of three replications. Each antibiotic mean that share the same lowercase letters are not significantly different; means with different letters are significantly different (p < 0.05). The error bars indicate standard deviation from the mean.
Figure 10.
Antibiotic sensitivity profile of select V. lutrae (i.e., VHT84 and VHT85). Each pair of lower/lower and lower/upper case letters represents a non-significant (p > 0.05) and significant (p < 0.05) difference in antibiotic sensitivity, respectively, among V. lutrae strains examined. Data bars (i.e., 48 h data only) represent the mean of three replications. Each antibiotic mean that share the same lowercase letters are not significantly different; means with different letters are significantly different (p < 0.05). The error bars indicate standard deviation from the mean.
Table 1.
List of raw seafood used in this study.
Table 1.
List of raw seafood used in this study.
| Seafood Type | Source | Sample # | Storage Condition |
|---|
| Crab | Beach | 2 | 4 °C |
| Oyster | Lagoon | 3 | 4 °C |
| Shrimp | Wet market | 3 | 4 °C |
Table 2.
Strains of V. lutrae (√VL) and non-V. lutrae (VP, JC, SE, VA, MN, VC, and VM) recovered in this study.
Table 2.
Strains of V. lutrae (√VL) and non-V. lutrae (VP, JC, SE, VA, MN, VC, and VM) recovered in this study.
Strain
ID 1 | 16S ID 3 | Colony Color | Nourishment Conditions 4 | Seafood Type |
|---|
| NaCl, pH | Time (h) | Step # |
|---|
| VHT1 | VP | G | M | 48 | 2 | Oyster |
| VHT2 | VP | G | M | 48 | 2 | Oyster |
| VHT5 | VL | Y | m | 48 | 2 | Crab |
| VHT6 | √VL | Y | m | 48 | 2 | Crab |
| VHT7 | √VL | Y | m | 48 | 2 | Shrimp |
| VHT9 | JC | G | E | 48 | 2 | Shrimp |
| VHT10 | JC | G | E | 48 | 2 | Shrimp |
| VHT11 | JC | G | E | 48 | 2 | Shrimp |
| VHT12 | JC | G | E | 48 | 2 | Shrimp |
| VHT13 | JC | G | E | 48 | 2 | Shrimp |
| VHT14 | VP | G | M | 48 | 2 | Oyster |
| VHT15 | VP | G | M | 48 | 2 | Oyster |
| VHT16 | VP | G | M | 48 | 2 | Oyster |
| VHT17 | VP | G | M | 24 | 1 | Oyster |
| VHT18 | VP | G | M | 24 | 1 | Oyster |
| VHT20 | VP | G | M | 24 | 1 | Oyster |
| VHT21 | VP | G | M | 24 | 1 | Oyster |
| VHT22 | VP | G | E | 72 | 1 | Oyster |
| VHT23 | SE | G | E | 72 | 1 | Oyster |
| VHT25 | VP | G | E | 72 | 1 | Oyster |
| VHT26 | VP | G | E | 72 | 1 | Oyster |
| VHT27 | √VL | Y | m | 48 | 2 | Shrimp |
| VHT28 | √VL | Y | m | 48 | 2 | Shrimp |
| VHT29 | √VL | Y | m | 48 | 2 | Shrimp |
| VHT31 | VA | Y | m | 8 | 2 | Oyster |
| VHT32 | VA | Y | m | 8 | 2 | Oyster |
| VHT33 | VA | Y | m | 8 | 2 | Oyster |
| VHT37 | MN | Y | m | 48 | 1 | Oyster |
| VHT38 | MN | Y | m | 48 | 1 | Oyster |
| VHT39 | VC | Y | m | 48 | 1 | Oyster |
| VHT40 | VC | Y | m | 48 | 1 | Oyster |
| VHT41 | √VL | Y | m | 24 | 1 | Shrimp |
| VHT42 | VM | Y | m | 24 | 1 | Shrimp |
| VHT43 | √VL | Y | m | 24 | 1 | Shrimp |
| VHT44 | √VL | Y | m | 24 | 1 | Shrimp |
| VHT45 | VM | Y | m | 24 | 1 | Shrimp |
| VHT46 | VA | Y | m | 12 | 1 | Crab |
| VHT47 | VA | Y | m | 12 | 1 | Crab |
| VHT48 | VA | Y | m | 12 | 1 | Crab |
| VHT49 | VA | Y | m | 12 | 1 | Crab |
| VHT50 | VA | Y | m | 12 | 1 | Crab |
| VHT82 2 | √VL | NA | NA | NA | NA | VHT27 |
| VHT83 2 | √VL | NA | NA | NA | NA | VHT28 |
| VHT84 2 | √VL | NA | NA | NA | NA | VHT29 |
| VHT85 2 | √VL | NA | NA | NA | NA | VHT44 |
Table 3.
Bacterial colony-forming profile of enriched seafood on TCBS agar at the post step 1 of the 2-step enrichment method 1.
Table 3.
Bacterial colony-forming profile of enriched seafood on TCBS agar at the post step 1 of the 2-step enrichment method 1.
| Enrichment Time (h) 2 | Seafood Type and Enrichment Only Conditions 3 |
|---|
| Crab | Oyster | Shrimp |
|---|
| Mild | Moderate | Extreme | Mild | Moderate | Extreme | Mild | Moderate | Extreme |
|---|
| 0 | X | X | X | X | X | X | X | X | X |
| 8 | X | X | X | X | X | X | X | X | X |
| 12 | √, Y | X | X | X | X | X | X | X | X |
| 24 | X | X | X | X | √, G | X | √, Y+ | X | X |
| 48 | X | X | X | √, Y | X | X | X | X | X |
| 72 | X | X | X | X | X | √, G | X | X | X |
Table 4.
Bacterial colony-forming profile of enriched seafood on TCBS agar at the post steps 1 and 2 of the 2-step enrichment method 1 followed by overnight cold storage.
Table 4.
Bacterial colony-forming profile of enriched seafood on TCBS agar at the post steps 1 and 2 of the 2-step enrichment method 1 followed by overnight cold storage.
| Enrichment Time (h) 2 | Seafood Type and Enrichment Conditions 3 + Heat |
|---|
| Crab | Oyster | Shrimp |
|---|
| Mild | Moderate | Extreme | Mild | Moderate | Extreme | Mild | Moderate | Extreme |
|---|
| 0 | X | X | X | X | X | X | X | X | X |
| 8 | X | X | X | √, Y | X | X | X | X | X |
| 12 | X | X | X | X | X | X | X | X | X |
| 24 | X | X | X | X | X | X | X | X | X |
| 48 | √, Y+ | X | X | X | √, G | X | √, Y+ | X | √, G |
| 72 | X | X | X | X | X | X | X | X | X |
Table 5.
Pasteurization sensitivity of heat-resistant V. lutrae isolated in this study.
Table 5.
Pasteurization sensitivity of heat-resistant V. lutrae isolated in this study.
| Strain ID | Heat-Resistant 1 | Pateurization-Resistant 2 | Re-ID 3 |
|---|
| VHT27 | v | v | VHT82 |
| VHT28 | v | v | VHT83 |
| VHT29 | v | v | VHT84 |
Table 6.
Thermal resuscitation of VBNC V. lutrae lab strains isolated in this study.
Table 6.
Thermal resuscitation of VBNC V. lutrae lab strains isolated in this study.
| Strain ID 1 | 8 h Thermal 2 | 1.5 h Thermal 2 | Heat-Resistant 3 | Regular 4 |
|---|
| VHT5 | - | - | v | NA |
| VHT6 | - | - | v | NA |
| VHT7 | - | ND | v | NA |
| VHT27 | ND | ND | v | NA |
| VHT28 | ND | - | v | NA |
| VHT29 | ND | - | v | NA |
| VHT41 | - | - | NA | v |
| VHT43 | - | - | NA | v |
| VHT44 | + | - | NA | v |
| VHT82 | ND | + | VHT27 der. | NA |
| VHT83 | ND | - | VHT28 der. | NA |
| VHT84 | ND | - | VHT29 der. | NA |
| VHT85 | ND | - | NA | VHT44 der. |
Table 7.
Oxygen-dependent growth profile of select V. lutrae isolates.
Table 7.
Oxygen-dependent growth profile of select V. lutrae isolates.
| O2 Availability 1 | V. lutrae Strains 2 in This Study |
|---|
| VHT28 | VHT29 | VHT82 | VHT83 | VHT84 | VHT85 |
|---|
| Layer (full) | ND | ND | v | v | x | v |
| Wrap (minute) | x | v | ND | x | v | v |
| Anaerobic jar (zero) | ND | ND | v | ND | ND | ND |
| Classification | Strict aerobe | Microaerobe | Facultative anaerobe | Strict aerobe | Microaerobe | Facultative anaerobe |
Table 8.
Antibiotic interpreted sensitivity 1 and MAR index of select V. lutrae strains.
Table 8.
Antibiotic interpreted sensitivity 1 and MAR index of select V. lutrae strains.
| Antibiotic | VHT84 | VHT85 |
|---|
| Avg Inhibition size (mm) ± S.D. | S | I | R | Avg Inhibition size (mm) ± S.D. | S | I | R |
|---|
| AMC30 | 40 ± 0.82 | v | | | 40 ± 1.25 | v | | |
| CEF30 | 40 ± 1.25 | v | | | 39 ± 0.82 | v | | |
| CLI2 | 38 ± 1.25 | v | | | 39 ± 0.82 | v | | |
| DOX30 | 40 ± 0.47 | v | | | 40 ± 0.82 | v | | |
| ERY15 | 38 ± 0.47 | v | | | 40 ± 0.47 | v | | |
| GEN10 | 13 ± 0.82 | | v | | 8 ± 0.47 | | | v |
| RIF5 | 41 ± 0.94 | v | | | 41 ± 0.47 | v | | |
| VAN30 | 29 ± 0.82 | v | | | 39 ± 0.82 | v | | |
| MAR index | | 0 | | 0.125 |