Role of the Mn-Catalase in Aerobic Growth of Lactobacillus plantarum ATCC 14431
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Measurements
2.2. Chemicals and Enzymes
2.3. Preparation of Cell-Free Extracts (CFEs)
2.4. Biochemical Assays
2.5. Western Blotting
2.6. Effects of H2O2 on Growth (OD600nm)
2.7. Microscopy and Imaging
3. Results
3.1. NC1543 Is Devoid of Catalase (MnKat)
3.2. MnKat and Cellular Physiology
3.2.1. Effect of MnKat on Biochemical Profile
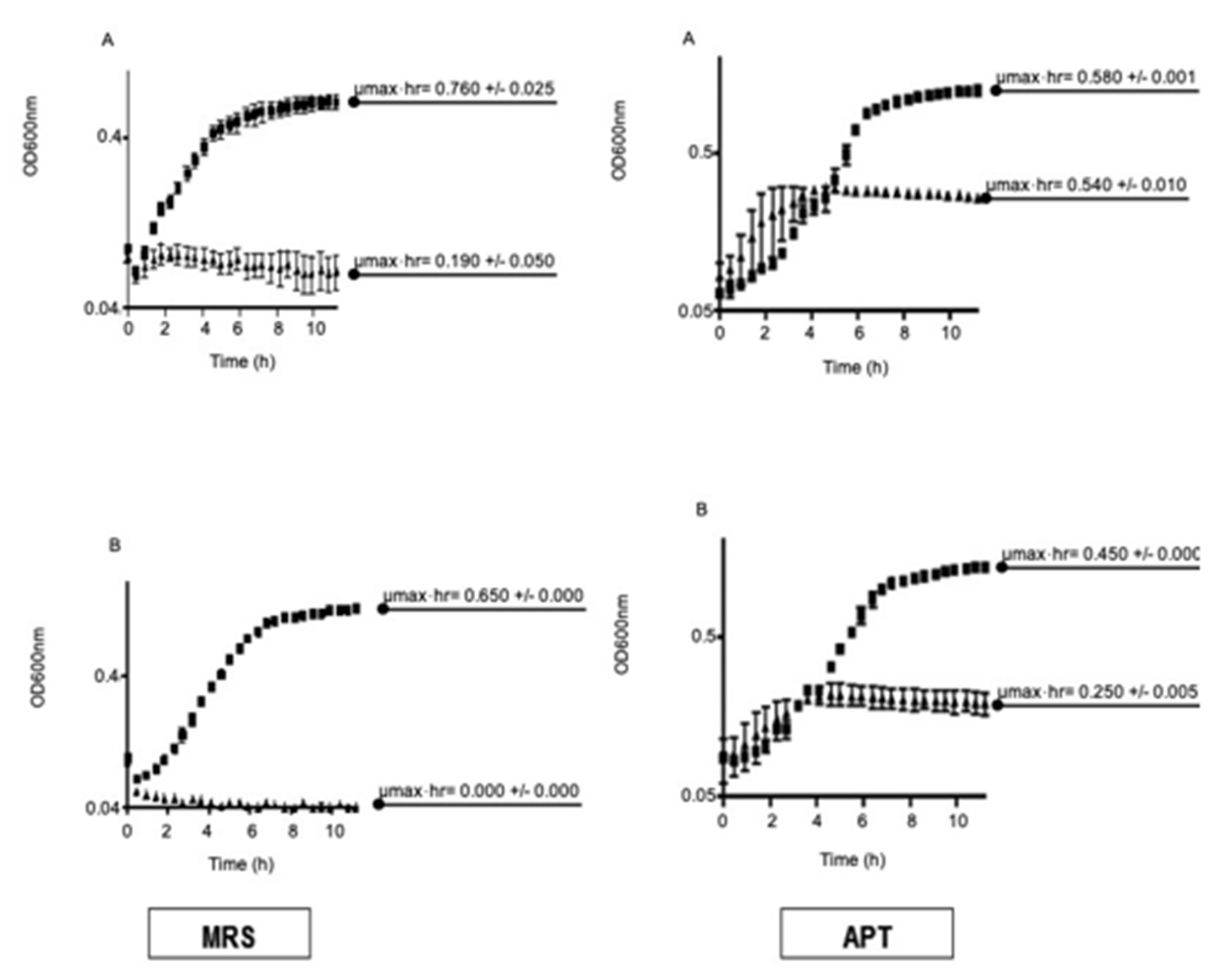
3.2.2. Effect of MnKat on Sensitivity to H2O2
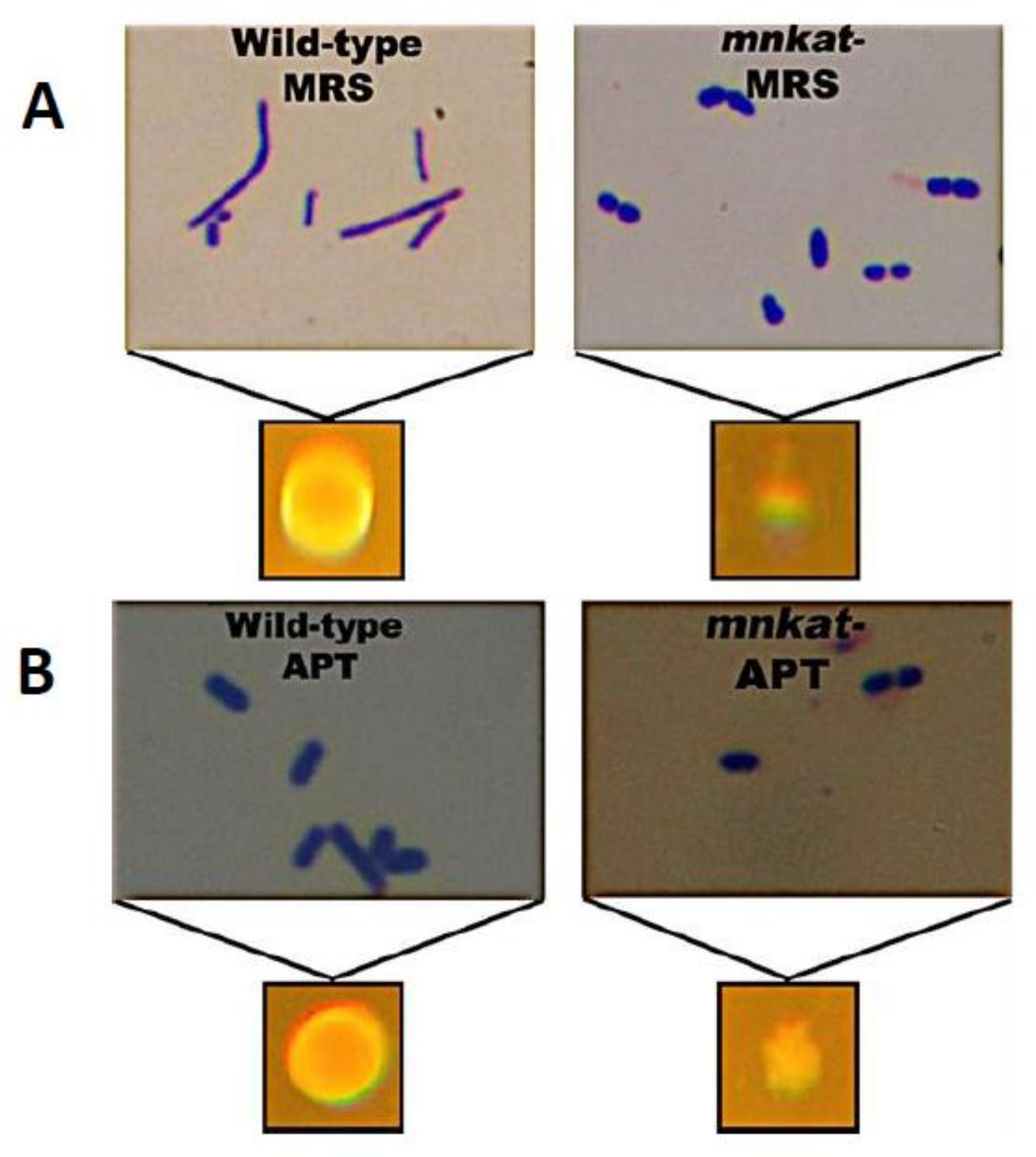
3.2.3. Effect of MnKat on Cell and Colony Morphology
4. Discussion
4.1. How the Absence of the MnKat ENZYME CAUSES the Observed Changes in Cell Morphology?
4.2. How can Manganese Protect against Hydrogen Peroxide Toxicity?
5. Conclusions
- Under aerobic conditions, MnKat is essential for normal aerobic growth of L. plantarum ATCC 14431; it is required for removing endogenous H2O2;
- Inactivation of MnKat results in hyper-sensitivity to added H2O2; and
- Media containing high Mn concentrations (e.g., APT) improves the growth of the MnKat mutant strain (NC1543) relative to that seen in the low Mn medium (e.g., MRS).
- This information will help to guide future investigations into alternative mechanisms for ROS detoxification and broaden studies on the role of Mn-catalases in other members of the LAB group as well as other prokaryotes. The findings will also bring awareness of the need to protect economically important organisms (e.g., LAB and probiotics) against oxidative stress, which is very important in the food fermentation and microbiome studies.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Archibald, F.S. Manganese: Its Acquistion by and Function in the Lactic Acid Bacteria. CRC Crit. Rev. Microbiol. 1986, 13, 63–109. [Google Scholar] [CrossRef]
- Condon, S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 1987, 46, 269–280. [Google Scholar] [CrossRef]
- Gotz, F.; Sedewitz, B.; Elstner, E.F. Oxygen utilization by Lactobacillus plantarum. I. Oxygen consuming reactions. Arch. Microbiol. 1980, 125, 209–214. [Google Scholar] [CrossRef]
- Murphy, M.G.; Condon, S. Correlation of oxygen utilization and hydrogen peroxide accumulation with oxygen induced enzymes in Lactobacillus plantarum cultures. Arch. Microbiol. 1984, 138, 44–48. [Google Scholar] [CrossRef]
- Teusink, B.; van Enckevort, F.H.; Francke, C.; Wiersma, A.; Wegkamp, A.; Smid, E.J.; Siezen, R.J. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: Comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 2005, 71, 7253–7262. [Google Scholar] [CrossRef]
- Archibald, F.S.; Duong, M.N. Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 1984, 158, 1–8. [Google Scholar] [CrossRef]
- Archibald, F.S.; Fridovich, I. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 1981, 145, 442–451. [Google Scholar] [CrossRef]
- Abriouel, H.; Herrmann, A.; Starke, J.; Yousif, N.M.; Wijaya, A.; Tauscher, B.; Holzapfel, W.; Franz, M. Cloning and heterologous expression of hematin-dependent catalase produced by Lactobacillus plantarum CNRZ 1228. Appl. Environ. Microbiol. 2004, 70, 603–606. [Google Scholar] [CrossRef]
- Amanatidou, A.; Smid, E.J.; Bennik, M.H.; Gorris, L.G. Antioxidative properties of Lactobacillus sake upon exposure to elevated oxygen concentrations. FEMS Microbiol. Lett. 2001, 203, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Andrus, J.M.; Bowen, S.W.; Klaenhammer, T.R.; Hassan, H.M. Molecular characterization and functional analysis of the manganese-containing superoxide dismutase gene (sodA) from Streptococcus thermophilus AO54. Arch. Biochem. Biophys. 2003, 420, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Archibald, F.S.; Fridovich, I. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J. Bacteriol. 1981, 146, 928–936. [Google Scholar] [CrossRef]
- Dacre, J.C.; Sharpe, M.E. Catalase production by Lactobacilli. Nature 1956, 178, 700. [Google Scholar] [CrossRef]
- De Angelis, M.; Gobbetti, M. Lactobacillus sanfranciscensis CB1: Manganese, oxygen, superoxide dismutase and metabolism. Appl. Microbiol. Biotechnol. 1999, 51, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Hertel, C.; Schmidt, G.; Fischer, M.; Oellers, K.; Hammes, W.P. Oxygen-dependent regulation of the expression of the catalase gene katA of Lactobacillus sakei LTH677. Appl. Environ. Microbiol. 1998, 64, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- Johnston, M.A.; Delwiche, E.A. Distribution and Characteristics of the catalases of Lactobacilliacaea. J. Bacteriol. 1965, 90, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Johnston, M.A.; Delwiche, E.A. Isolation and Characterization of the cyanide-resistant and azide-resistant catalase of Lactobacillus plantarum. J. Bacteriol. 1965, 90, 352–356. [Google Scholar] [CrossRef]
- Knauf, H.J.; Vogel, R.F.; Hammes, W.P. Cloning, sequence, and phenotypic expression of katA, which encodes the catalase of Lactobacillus sake LTH677. Appl. Environ. Microbiol. 1992, 58, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Kono, Y.; Fridovich, I. Isolation and characterization of the pseudocatalase of Lactobacillus plantarum. J. Biol. Chem. 1983, 258, 6015–6019, Accession Number: 6853475. [Google Scholar] [CrossRef]
- Kullisaar, T.; Zilmer, M.; Mikelsaar, M.; Vihalemm, T.; Annuk, H.; Kairane, C.; Kilk, A. Two antioxidative lactobacilli strains as promising probiotics. Int. J. Food Microbiol. 2002, 72, 215–224. [Google Scholar] [CrossRef]
- Noonpakdee, W.; Sitthimonchai, S.; Panyim, S.; Lertsiri, S. Expression of the catalase gene katA in starter culture Lactobacillus plantarum TISTR850 tolerates oxidative stress and reduces lipid oxidation in fermented meat product. Int. J. Food Microbiol. 2004, 95, 127–135. [Google Scholar] [CrossRef]
- Poyart, C.; Berche, P.; Trieu-Cuot, P. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol. Lett. 1995, 131, 41–45. [Google Scholar] [CrossRef][Green Version]
- Sanders, J.W.; Leenhouts, K.J.A.; Haandrikman, J.; Venema, G.; Kok, J. Stress response in Lactococcus lactis: Cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J. Bacteriol. 1995, 177, 5254–5260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wolf, G.; Strahl, A.; Meisel, J.; Hammes, W.P. Heme-dependent catalase activity of lactobacilli. Int. J. Food Microbiol. 1991, 12, 133–140. [Google Scholar] [CrossRef]
- Hammes, W.P.; Hertel, C. The Genera Lactobacillus and Carnobacterium. Prokaryotes 2006, 4, 320–403. [Google Scholar]
- Igarashi, T.; Kono, Y.; Tanaka, K. Molecular cloning of manganese catalase from Lactobacillus plantarum. J. Biol. Chem. 1996, 271, 29521–29524. [Google Scholar] [CrossRef]
- Rochat, T.; Gratadoux, J.J.; Gruss, A.; Corthier, G.; Maguin, E.; Langella, P.; van de Guchte, M. Production of a heterologous nonheme catalase by Lactobacillus casei: An efficient tool for removal of H2O2 and protection of Lactobacillus bulgaricus from oxidative stress in milk. Appl. Environ. Microbiol. 2006, 72, 5143–5149. [Google Scholar] [CrossRef]
- Barynin, V.V.; Whittaker, M.M.; Antonyuk, S.V.; Lamzin, V.S.; Harrison, P.M.; Artymiuk, P.J.; Whittaker, J.W. Crystal structure of manganese catalase from Lactobacillus plantarum. Structure 2001, 9, 725–738. [Google Scholar] [CrossRef]
- Whittaker, M.M.; Barynin, V.V.; Igarashi, T.; Whittaker, J.W. Outer sphere mutagenesis of Lactobacillus plantarum manganese catalase disrupts the cluster core: Mechanistic implications. Eur. J. Biochem. 2003, 270, 1102–1116. [Google Scholar] [CrossRef]
- Kono, Y.; Fridovich, I. Functional significance of manganese catalase in Lactobacillus plantarum. J. Bacteriol. 1983, 155, 742–746. [Google Scholar] [CrossRef]
- Peacock, T.J. Biological role of Mn-catalase in select Lactobacilli. Ph.D. Thesis, North Carolina State Unviersity, Raleigh, NC, USA, 2008. Available online: http://www.lib.ncsu.edu/resolver/1840.16/4007 (accessed on 10 June 2021).
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Clare, D.A.; Duong, M.N.; Darr, D.; Archibald, F.; Fridovich, I. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 1984, 140, 532–537. [Google Scholar] [CrossRef]
- Beers, R.F.J.; Sizer, I.W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952, 195, 133–140, Accession Number: 14938361. [Google Scholar] [CrossRef]
- Claus, G.W. Understanding Microbes: A Laboratory Textbook for Microbiology; W.H. Freeman & Co.: New York, NY, USA, 1989. [Google Scholar]
- Hassan, H.M. Oxygen toxicity and mutagenicity in procaryotes. In Oxy Radicals and their Schavengers Systems; Molecular Aspects. Cohen, G., Greenwald, R., Eds.; Elsevier: Amsterdam, The Netherlands, 1983; Volume 1, pp. 198–206. [Google Scholar]
- Zuljan, F.A.; Repizo, G.D.; Alarcon, S.H.; Magni, C. α-Acetolactate synthase of Lactococcus lactis contributes to pH homeostasis in acid stress conditions. Int. J. Food Microbiol. 2014, 188, 99–107. [Google Scholar] [CrossRef]
- Liu, S. A Simple method to generate chromosomal mutations in Lactobacillus plantarum strain TF103 to eliminate undesired fermentation products. Appl. Biochem. Biotechnol. 2006, 131, 854–863. [Google Scholar] [CrossRef]
- Schellhorn, H.; Hassan, H.M. Response of hydroperoxidase and superoxide dismutase deficient mutants of Escherichia coli to oxidative stress. Can. J. Microbiol. 1988, 34, 1171–1176. [Google Scholar] [CrossRef]
- Pérez-Núňez, D.; Briandet, R.; David, B.; Gautier, C.; Renault, P.; Hallet, B.; Hols, P.; Carballido-López, R.; Guédon, E. A new morphogenesis pathway in bacteria: Unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol. Microbiol. 2011, 79, 759–771. [Google Scholar] [CrossRef]
- Bendezu, F.O.; de Boer, P.A. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J. Bacteriol. 2008, 190, 1792–1811. [Google Scholar] [CrossRef]
- Den Blaauwen, T.; de Pedro, M.A.; Nguyen-Disteche, M.; Ayala, J.A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 2008, 32, 321–344. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Montgomery, B.L. Reactive oxygen species are involved in the morphology-determining mechanism of Fremyella diplosiphon cells during complementary chromatic adaptation. Microbiology 2012, 158, 2235–2245. [Google Scholar] [CrossRef]
- Molina-Hoppner, A.; Sato, T.; Kato, C.; Ganzle, M.G.; Vogel, R.F. Effects of pressure on cell morphology and cell division of lactic acid bacteria. Extremophiles 2003, 7, 511–516. [Google Scholar] [CrossRef]
- Thibessard, A.; Fernandez, A.; Gintz, B.; Leblond-Bourget, N.; Decaris, B. Effects of rodA and pbp2b disruption on cell morphology and oxidative stress response of Streptococcus thermophiles CNRZ368. J. Bacteriol. 2002, 184, 2821–2826. [Google Scholar] [CrossRef]
- Jankovic, I.; Ventura, M.; Meylan, V.; Rouvet, M.; Elli, M.; Zink, R. Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillus gasseri 4B2. J. Bacteriol. 2003, 185, 3288–3296. [Google Scholar] [CrossRef]
- Macleod, R.A.; Snell, E.E. Some mineral requirements of the lactic acid bacteria. J. Biol. Chem. 1947, 170, 351–365. [Google Scholar] [CrossRef]
- Nierop Groot, M.N.; de Bont, J.A. Involvement of manganese in conversion of phenylalanine to benzaldehyde by lactic acid bacteria. Appl. Environ. Microbiol. 1999, 65, 5590–5593. [Google Scholar] [CrossRef]
- Nierop Groot, M.N.; de Bont, J.A.M. Conversion of phenylalanine to benzaldehyde initiated by an aminotransferase in Lactobacillus plantarum. Appl. Environ. Microbiol. 1998, 64, 3009–3013. [Google Scholar] [CrossRef]
- Raccach, M.; Marshall, P.S. Effect of manganese ions on the fermentative activity of frozen-thawed lactobacilli. J. Food Sci. 1985, 50, 665–668. [Google Scholar] [CrossRef]
- Stetter, K.O.; Zillig, W. Transcription in lactobacillaceae. DNA-dependent RNA polymerase from Lactobacillus curvatus. Eur. J. Biochem. 1974, 48, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Groot, M.N.; Klaassens, E.; de Vos, W.M.; Delcour, J.; Hols, P.; Kleerebezem, M. Genome-based in silico detection of putative manganese transport systems in Lactobacillus plantarum and their genetic analysis. Microbiology 2005, 151, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Chen, S.; Wilson, D.B. Cloning, expression, and characterization of cadmium and manganese uptake genes from Lactobacillus plantarum. Appl. Environ. Microbiol. 1999, 65, 4746–4752. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Reiske, H.R.; Wilson, D.B. Characterization of cadmium uptake in Lactobacillus plantarum and isolation of cadmium and manganese uptake mutants. Appl. Environ. Microbiol. 1999, 65, 4741–4745. [Google Scholar] [CrossRef] [PubMed]
- Archibald, F.S.; Fridovich, I. Investigations of the state of manganese in Lactobacillus plantarum. Arch. Biochem. Biophys. 1982, 215, 589–596. [Google Scholar] [CrossRef]
- Cheton, P.L.; Archibald, F.S. Manganese complexes and the generation and scavenging of hydroxyl free radicals. Free. Rad. Biol. Med. 1988, 5, 325–333. [Google Scholar] [CrossRef]
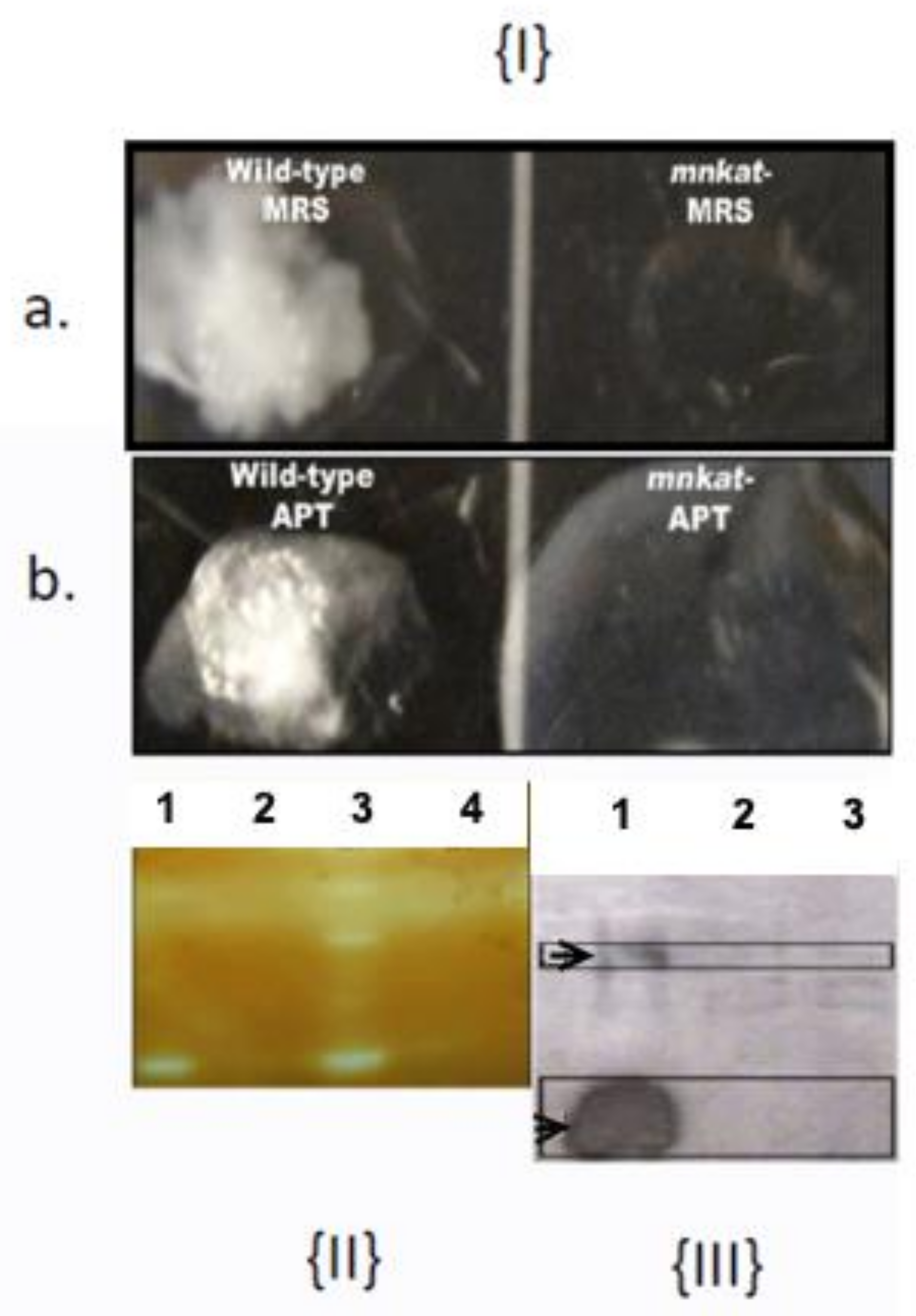

| Tests a | Reactions/Enzymes | Aerobic | Anaerobic | ||
|---|---|---|---|---|---|
| WT | Mutant | WT | Mutant | ||
| ONPG | β-galactosidase | − | − | − | − |
| ADH | Arginine dehydrolase | − | − | − | − |
| LDC | Lysine decarboxylase | − | − | − | − |
| ODC | Ornithine decarboxylase | − | − | − | − |
| CIT | Citrate utilization | − | − | − | − |
| H2S | Hydrogen Sulfide production | − | − | − | − |
| URE | Urease | − | − | − | − |
| TDA | Tryptophan deaminase | − | − | − | − |
| IND | Indole | − | − | − | − |
| VP | Vogues-Proskauer | + | weak b | + | + |
| GEL | Gelatin liquefaction | − | − | − | − |
| GLU | Glucose utilization | + | + | + | + |
| MAN | Mannitol utilization | + | + | + | + |
| INO | Inositol utilization | + | + | + | + |
| SOR | Sorbitol utilization | + | + | + | + |
| RHA | Rhamnose utilization | + | + | + | + |
| SAC | Sucrose utilization | + | + | + | + |
| MEL | Melibiose utilization | + | + | + | + |
| AMY | Amygdalin utilization | + | + | + | + |
| ARA | Arabinose utilization | + | + | + | + |
| NIT RED | Nitrate reductase | + | + | + | + |
| CAT | Catalase | +c | − | + | −d |
| OX | Oxidase | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peacock, T.; Hassan, H.M. Role of the Mn-Catalase in Aerobic Growth of Lactobacillus plantarum ATCC 14431. Appl. Microbiol. 2021, 1, 615-625. https://doi.org/10.3390/applmicrobiol1030040
Peacock T, Hassan HM. Role of the Mn-Catalase in Aerobic Growth of Lactobacillus plantarum ATCC 14431. Applied Microbiology. 2021; 1(3):615-625. https://doi.org/10.3390/applmicrobiol1030040
Chicago/Turabian StylePeacock, Trent, and Hosni M. Hassan. 2021. "Role of the Mn-Catalase in Aerobic Growth of Lactobacillus plantarum ATCC 14431" Applied Microbiology 1, no. 3: 615-625. https://doi.org/10.3390/applmicrobiol1030040
APA StylePeacock, T., & Hassan, H. M. (2021). Role of the Mn-Catalase in Aerobic Growth of Lactobacillus plantarum ATCC 14431. Applied Microbiology, 1(3), 615-625. https://doi.org/10.3390/applmicrobiol1030040







