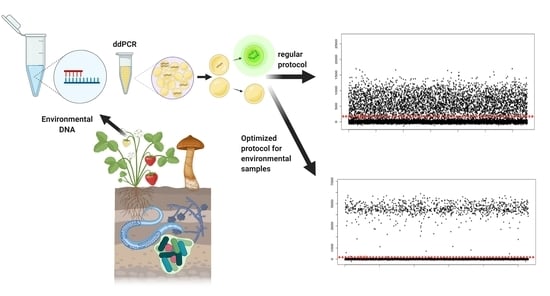
Challenges Using Droplet Digital PCR for Environmental Samples
Abstract
1. Introduction
2. Materials and Methods
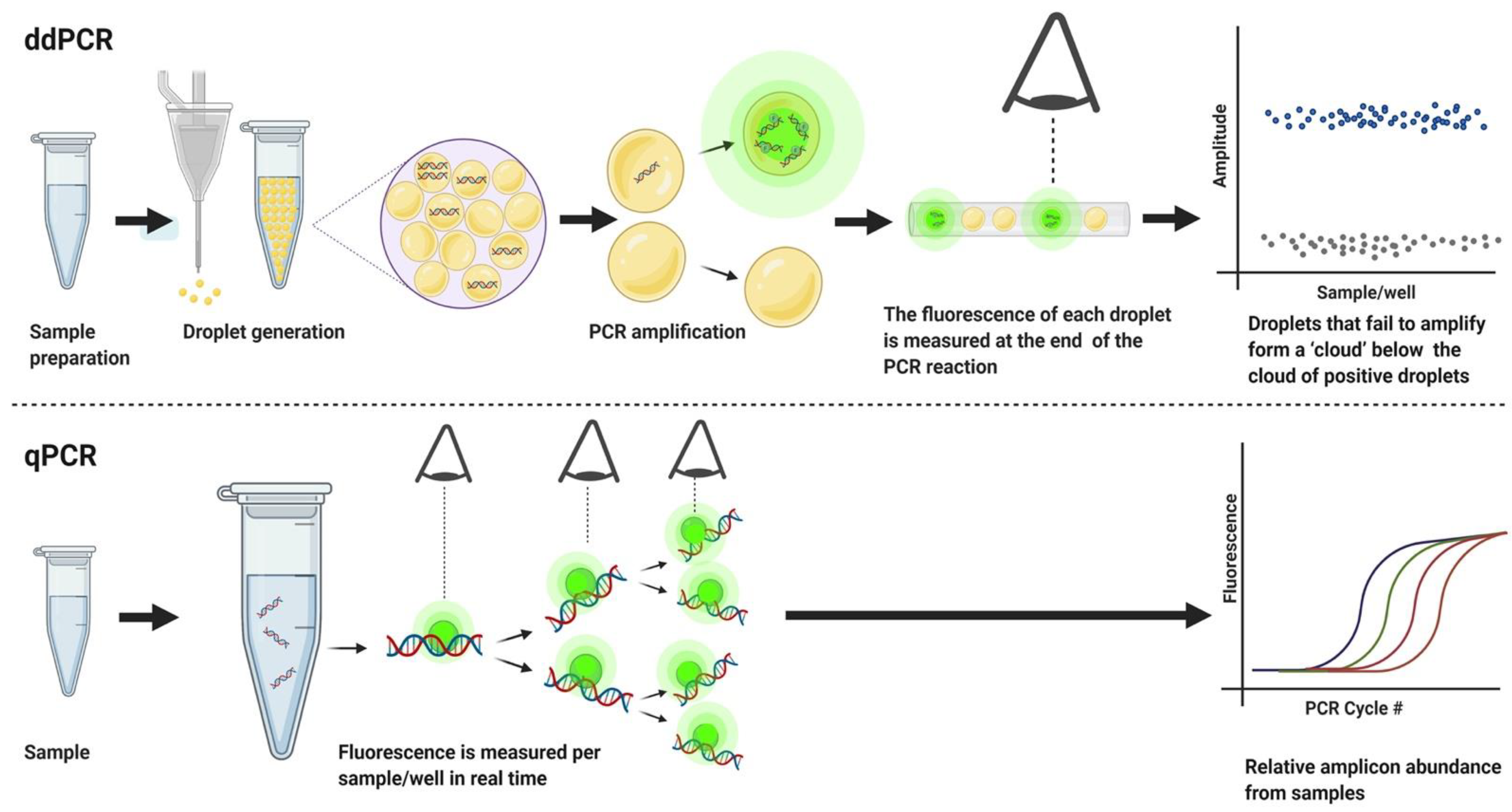
2.1. Hardware and Software
2.2. Origin of Environmental DNA Used to Optimize ddPCR
2.3. DNA Extraction and Primers/Probe
- Forward: 5′- AGCAAATCTAAGTTCCTCAGAG -3′
- Reverse: 5′- ACTTCTATGGCTTTGTACAGG-3′
- Probe: FAM/CCCACCAGG/ZEN/GCAGATTAATCTTCCTT/3IABKFQ-3
- Standardized cycling conditions and ddPCR reaction mixture
2.4. Parameters Tested
2.4.1. Primer/Probe Concentration
2.4.2. Sample DNA Concentration
2.4.3. Automated Algorithm Comparison
3. Results
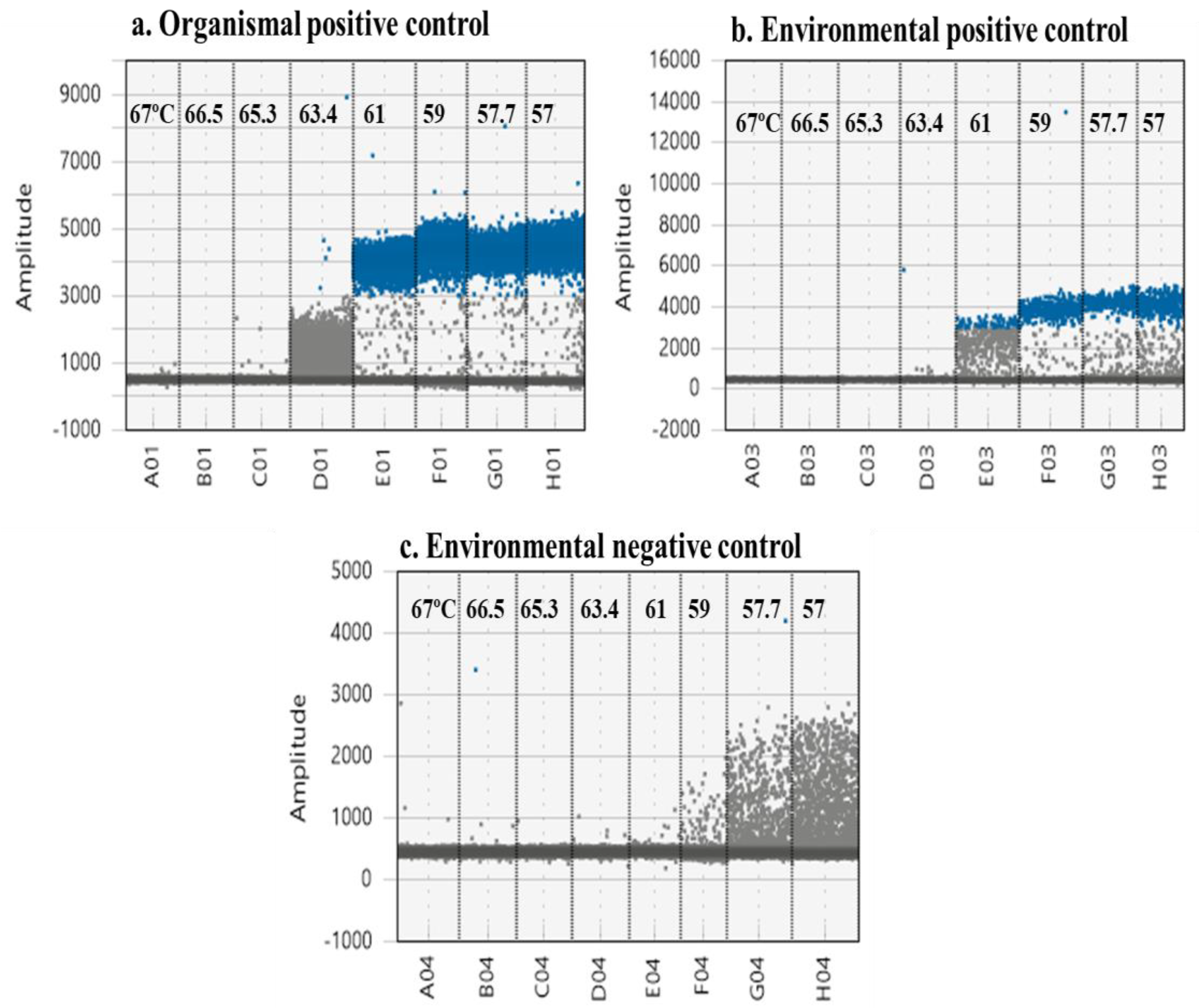
3.1. Optimizing Cycling Conditions
3.2. Optimizing Annealing/Extension Temperature
3.3. Adjusting Primer/Probe Concentration
3.4. Adjusting Sample Concentration
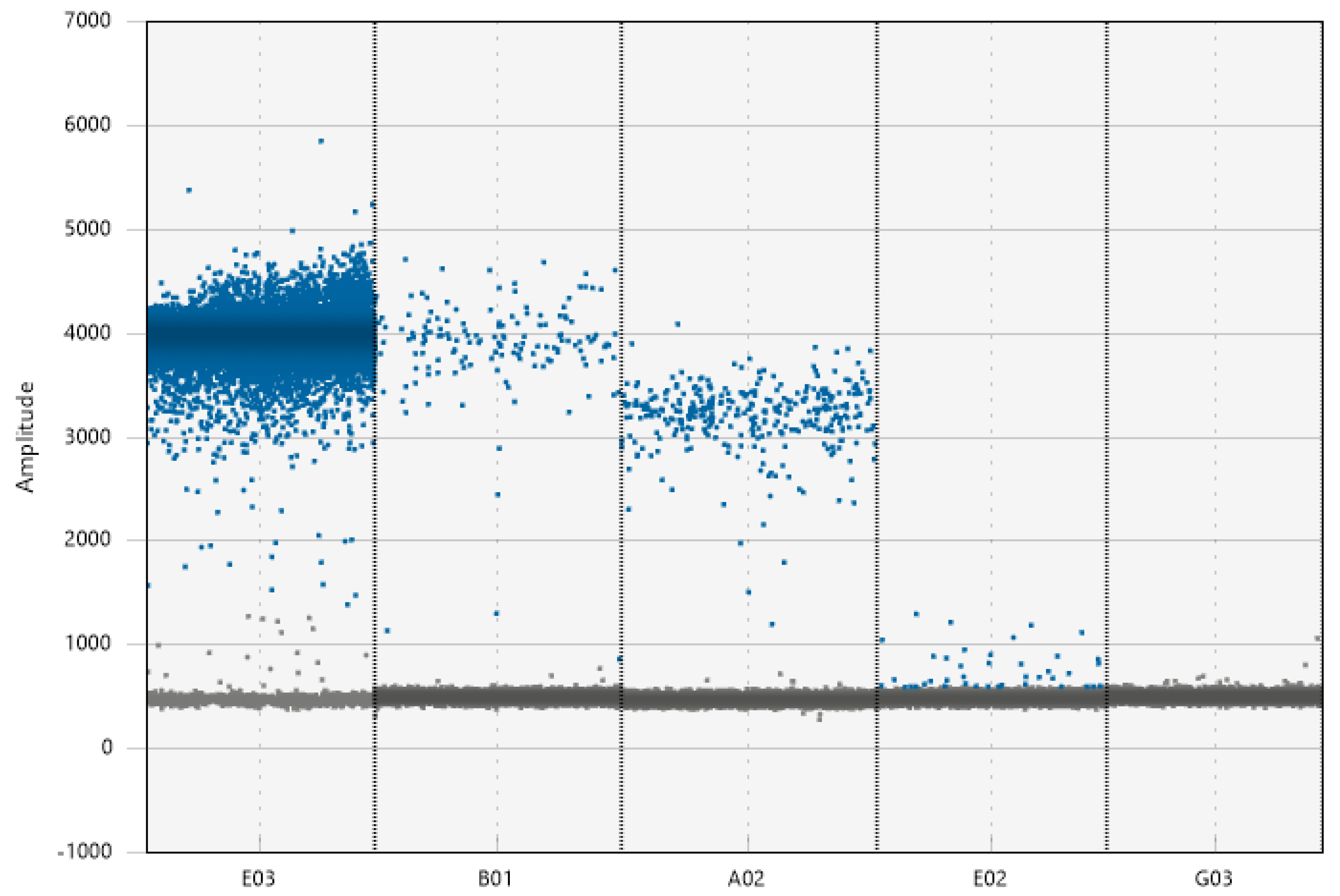
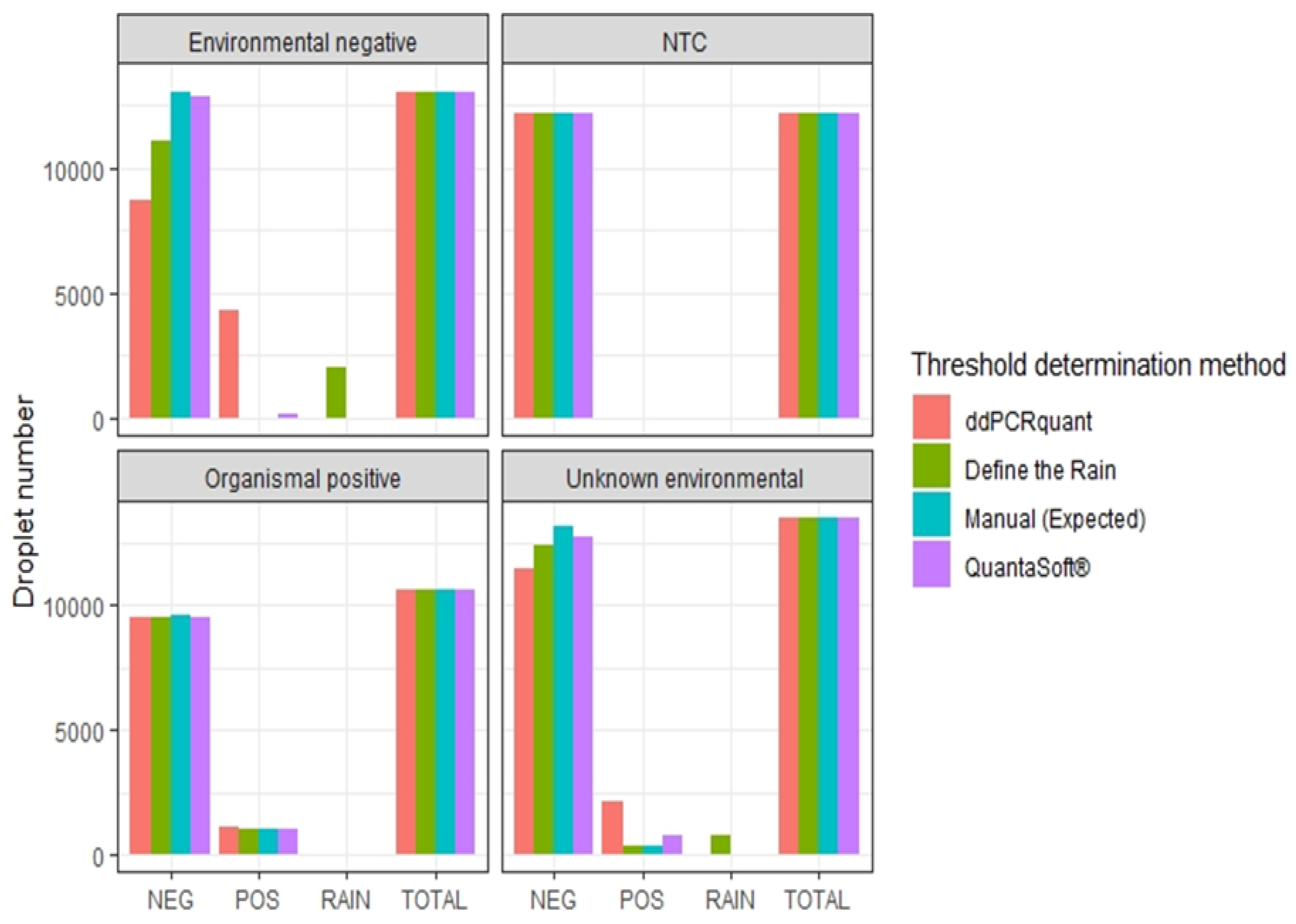
3.5. Threshold Determination
4. Discussion
4.1. Cycling Conditions
4.2. Primer/Probe Concentration
4.3. Sample Concentration
4.4. Threshold Determination
4.5. Limit of Detection Determination
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.S. Digital Assays Part I: Partitioning Statistics and Digital PCR. SLAS Technol. Transl. Life Sci. Innov. 2017, 22, 369–386. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.L.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of Cancer DNA in Plasma of Patients with Early-Stage Breast Cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhao, X.; Guo, W.; Huang, M.; Qiu, L.; Zhang, W.; Zhang, Z.; Li, W.; Zhu, X.; Chen, Z. Dynamic monitoring of HER2 amplification in circulating DNA of patients with metastatic colorectal cancer treated with cetuximab. Clin. Transl. Oncol. 2019, 22, 928–934. [Google Scholar] [CrossRef]
- Fu, W.; Wang, C.; Zhu, P.; Xu, W.; Li, X.; Zhu, S. A universal analytical approach for screening and monitoring of authorized and unauthorized GMOs. LWT 2020, 125, 109176. [Google Scholar] [CrossRef]
- Milavec, M.; Dobnik, D.; Yang, L.; Zhang, D.; Gruden, K.; Žel, J. GMO quantification: Valuable experience and insights for the future. Anal. Bioanal. Chem. 2014, 406, 6485–6497. [Google Scholar] [CrossRef]
- Hamza, I.A.; Bibby, K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods 2019, 266, 11–24. [Google Scholar] [CrossRef] [PubMed]
- NejcRački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 1–10. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Yang, R.; Paparini, A.; Monis, P.; Ryan, U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 2014, 44, 1105–1113. [Google Scholar] [CrossRef]
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef]
- Trypsteen, W.; Vynck, M.; De Neve, J.; Bonczkowski, P.; Kiselinova, M.; Malatinkova, E.; Vervisch, K.; Thas, O.; Vandekerckhove, L.; De Spiegelaere, W. ddpcRquant: Threshold determination for single channel droplet digital PCR experiments. Anal. Bioanal. Chem. 2015, 407, 5827–5834. [Google Scholar] [CrossRef] [PubMed]
- Zink, F.A.; Tembrock, L.R.; Timm, A.E.; Farris, R.E.; Perera, O.P.; Gilligan, T.M. A droplet digital PCR (ddPCR) assay to detect Helicoverpa armigera (Lepidoptera: Noctuidae) in bulk trap samples. PLoS ONE 2017, 12, e0178704. [Google Scholar] [CrossRef]
- Dobnik, D.; Demšar, T.; Huber, I.; Gerdes, L.; Broeders, S.; Roosens, N.; Debode, F.; Berben, G.; Žel, J. Inter-laboratory analysis of selected genetically modified plant reference materials with digital PCR. Anal. Bioanal. Chem. 2017, 410, 211–221. [Google Scholar] [CrossRef]
- Pipan, B.; Zupančič, M.; Blatnik, E.; Dolničar, P.; Meglič, V. Comparison of six genomic DNA extraction methods for molecular downstream applications of apple tree (Malus X domestica). Cogent Food Agric. 2018, 4. [Google Scholar] [CrossRef]
- Meijerink, J.; Mandigers, C.; Van De Locht, L.; Tönnissen, E.; Goodsaid, F.; Raemaekers, J. A Novel Method to Compensate for Different Amplification Efficiencies between Patient DNA Samples in Quantitative Real-Time PCR. J. Mol. Diagn. 2001, 3, 55–61. [Google Scholar] [CrossRef][Green Version]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 1–23. [Google Scholar] [CrossRef]
- Kreader, C.A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996, 62, 1102–1106. [Google Scholar] [CrossRef]
- Dingle, T.; Sedlak, R.H.; Cook, L.; Jerome, K.R. Tolerance of Droplet-Digital PCR vs. Real-Time Quantitative PCR to Inhibitory Substances. Clin. Chem. 2013, 59, 1670–1672. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Cavé, L.; Brothier, E.; Abrouk, D.; Bouda, P.S.; Hien, E.; Nazaret, S. Efficiency and sensitivity of the digital droplet PCR for the quantification of antibiotic resistance genes in soils and organic residues. Appl. Microbiol. Biotechnol. 2016, 100, 10597–10608. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kokkoris, V.; Li, Y.; Hamel, C.; Hanson, K.; Hart, M. Site specificity in establishment of a commercial arbuscular mycorrhizal fungal inoculant. Sci. Total. Environ. 2019, 660, 1135–1143. [Google Scholar] [CrossRef]
- Sieverding, E.; Da Silva, G.A.; Berndt, R.; Oehl, F. Rhizoglomus, a new genus of the Glomeraceae. Mycotaxon 2015, 129, 373–386. [Google Scholar] [CrossRef]
- Doi, H.; Uchii, K.; Takahara, T.; Matsuhashi, S.; Yamanaka, H.; Minamoto, T. Use of Droplet Digital PCR for Estimation of Fish Abundance and Biomass in Environmental DNA Surveys. PLoS ONE 2015, 10, e0122763. [Google Scholar] [CrossRef] [PubMed]
- Simmons, M.; Tucker, A.; Chadderton, W.L.; Jerde, C.L.; Mahon, A.R. Active and passive environmental DNA surveillance of aquatic invasive species. Can. J. Fish. Aquat. Sci. 2016, 73, 76–83. [Google Scholar] [CrossRef]
- Cooley, M.B.; Carychao, D.; Gorski, L. Optimized Co-extraction and Quantification of DNA from Enteric Pathogens in Surface Water Samples Near Produce Fields in California. Front. Microbiol. 2018, 9, 448. [Google Scholar] [CrossRef]
- del Pilar Martínez-Diz, M.; Andrés-Sodupe, M.; Berbegal, M.; Bujanda, M.R.; Díaz-Losada, E.; Gramaje, D. Droplet Digital PCR Technology for Detection ofIlyonectria liriodendrifrom Grapevine Environmental Samples. Plant Dis. 2020, 104, 1144–1150. [Google Scholar] [CrossRef]
- Rosa, D.; Pogiatzis, A.; Bowen, P.; Kokkoris, V.; Richards, A.; Holland, T.; Hart, M. Performance and Establishment of a Commercial Mycorrhizal Inoculant in Viticulture. Agriculture 2020, 10, 539. [Google Scholar] [CrossRef]
- Kokkoris, V.; Chagnon, P.-L.; Yildirir, G.; Clarke, K.; Goh, D.; MacLean, A.M.; Dettman, J.; Stefani, F.; Corradi, N. Host identity influences nuclear dynamics in arbuscular mycorrhizal fungi. Curr. Biol. 2021, 31, 1531–1538.e6. [Google Scholar] [CrossRef]
- McMahon, T.C.; Blais, B.W.; Wong, A.; Carrillo, C.D. Multiplexed Single Intact Cell Droplet Digital PCR (MuSIC ddPCR) Method for Specific Detection of Enterohemorrhagic E. coli (EHEC) in Food Enrichment Cultures. Front. Microbiol. 2017, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Williams, J.; Gärtner, K.; Phillips, R.; Hurst, J.; Frater, J. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, ‘definetherain’. J. Virol. Methods 2014, 202, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Dreo, T.; Pirc, M.; Živa, R.; Pavšič, J.; Milavec, M.; Žel, J.; Gruden, K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014, 406, 6513–6528. [Google Scholar] [CrossRef]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly Precise Measurement of HIV DNA by Droplet Digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef]
- Jacobs, B.K.M.; Goetghebeur, E.; Vandesompele, J.; De Ganck, A.; Nijs, N.; Beckers, A.; Papazova, N.; Roosens, N.H.; Clement, L. Model-Based Classification for Digital PCR: Your Umbrella for Rain. Anal. Chem. 2017, 89, 4461–4467. [Google Scholar] [CrossRef] [PubMed]
- Witte, A.K.; Mester, P.; Fister, S.; Witte, M.; Schoder, D.; Rossmanith, P. A Systematic Investigation of Parameters Influencing Droplet Rain in the Listeria monocytogenes prfA Assay—Reduction of Ambiguous Results in ddPCR. PLoS ONE 2016, 11, e0168179. [Google Scholar] [CrossRef] [PubMed]
- Köppel, R.; Bucher, T. Rapid establishment of droplet digital PCR for quantitative GMO analysis. Eur. Food Res. Technol. 2015, 241, 427–439. [Google Scholar] [CrossRef]
- Witte, A.K.; Fister, S.; Mester, P.; Schoder, D.; Rossmanith, P. Evaluation of the performance of quantitative detection of the Listeria monocytogenes prfA locus with droplet digital PCR. Anal. Bioanal. Chem. 2016, 408, 7583–7593. [Google Scholar] [CrossRef]
- Hunter, M.E.; Dorazio, R.M.; Butterfield, J.S.S.; Meigs-Friend, G.; Nico, L.G.; Ferrante, J.A. Detection limits of quantitative and digital PCR assays and their influence in presence-absence surveys of environmental DNA. Mol. Ecol. Resour. 2017, 17, 221–229. [Google Scholar] [CrossRef]
- Kiselinova, M.; Pasternak, A.O.; De Spiegelaere, W.; Vogelaers, D.; Berkhout, B.; Vandekerckhove, L. Comparison of Droplet Digital PCR and Seminested Real-Time PCR for Quantification of Cell-Associated HIV-1 RNA. PLoS ONE 2014, 9, e85999. [Google Scholar] [CrossRef]
- Baker, C.S.; Steel, D.; Nieukirk, S.; Klinck, H. Environmental DNA (eDNA) From the Wake of the Whales: Droplet Digital PCR for Detection and Species Identification. Front. Mar. Sci. 2018, 5. [Google Scholar] [CrossRef]



| Variable | Recommendations | Effect | |
|---|---|---|---|
| Optimizing cycling conditions | Denaturation | Increase to 1 min | Results are additive and can improve cluster separation as well as reduce rain between the clusters |
| Annealing/extension | Increase to 2 min | ||
| Ramp rate | Decrease to 1 °C/sec | ||
| Number of cycles | Increase to 41–45 | ||
| Optimizing annealing/extension temperature | Annealing/extension temperature | Using organismal positive control | Ensure the effectiveness of the assay |
| Use a high abundance positive environmental sample | Reveals the optimum temperature for increased fluorescence and less rain between clouds | ||
| Use a low in abundance positive environmental sample | Ensures the optimum temperature in which low abundance target is clearly separated from rain or non-specific amplification | ||
| Use a negative environmental sample | Reveals the temperature in which non-specific amplification is limited/absent reducing false positive recognition | ||
| Use a non-template control (NTC) | Reject the possibility of contamination | ||
| Adjusting primer/probe concentration | Primer/probe concentration | Use recommended concentration | Decreasing primer/probe quantity can reduce cluster separation by decreasing fluorescence levels. Increasing the concentration beyond recommended does not alter the fluorescence levels |
| Adjusting sample concentration | Sample concentration | Dilute very concentrated samples as needed when there is no clear cluster separation or a lot of rain between clusters | Identify false positives and avoid false negatives |
| Threshold determination | Manually set threshold (should be determined taking under consideration multiple controls) | Using a high abundance positive environmental sample | When the target is abundant fluorescence levels can be reduced. In case a high threshold has been chosen then it should not exceed the lower limits of that samples positive cloud |
| Using a negative environmental sample | Can help eliminate non-specific amplification by setting the threshold above the non-specific amplified cluster | ||
| Minimum accepted positive droplet threshold | Use three–five NTC samples. Use both a positive control dilution series and a spiked environmental sample with same dilution series | Determining the limit of detection can eliminate false positives due to instrument artefacts or fluorescence of foreign particles |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkoris, V.; Vukicevich, E.; Richards, A.; Thomsen, C.; Hart, M.M. Challenges Using Droplet Digital PCR for Environmental Samples. Appl. Microbiol. 2021, 1, 74-88. https://doi.org/10.3390/applmicrobiol1010007
Kokkoris V, Vukicevich E, Richards A, Thomsen C, Hart MM. Challenges Using Droplet Digital PCR for Environmental Samples. Applied Microbiology. 2021; 1(1):74-88. https://doi.org/10.3390/applmicrobiol1010007
Chicago/Turabian StyleKokkoris, Vasilis, Eric Vukicevich, Andrew Richards, Corrina Thomsen, and Miranda M. Hart. 2021. "Challenges Using Droplet Digital PCR for Environmental Samples" Applied Microbiology 1, no. 1: 74-88. https://doi.org/10.3390/applmicrobiol1010007
APA StyleKokkoris, V., Vukicevich, E., Richards, A., Thomsen, C., & Hart, M. M. (2021). Challenges Using Droplet Digital PCR for Environmental Samples. Applied Microbiology, 1(1), 74-88. https://doi.org/10.3390/applmicrobiol1010007