Towards Streamlining the Choice of Crossing Combinations in Plant Breeding by Integrating Model-Based Recommendations and Plant Breeder’s Preferences
Abstract
:1. Introduction
2. Materials and Methods
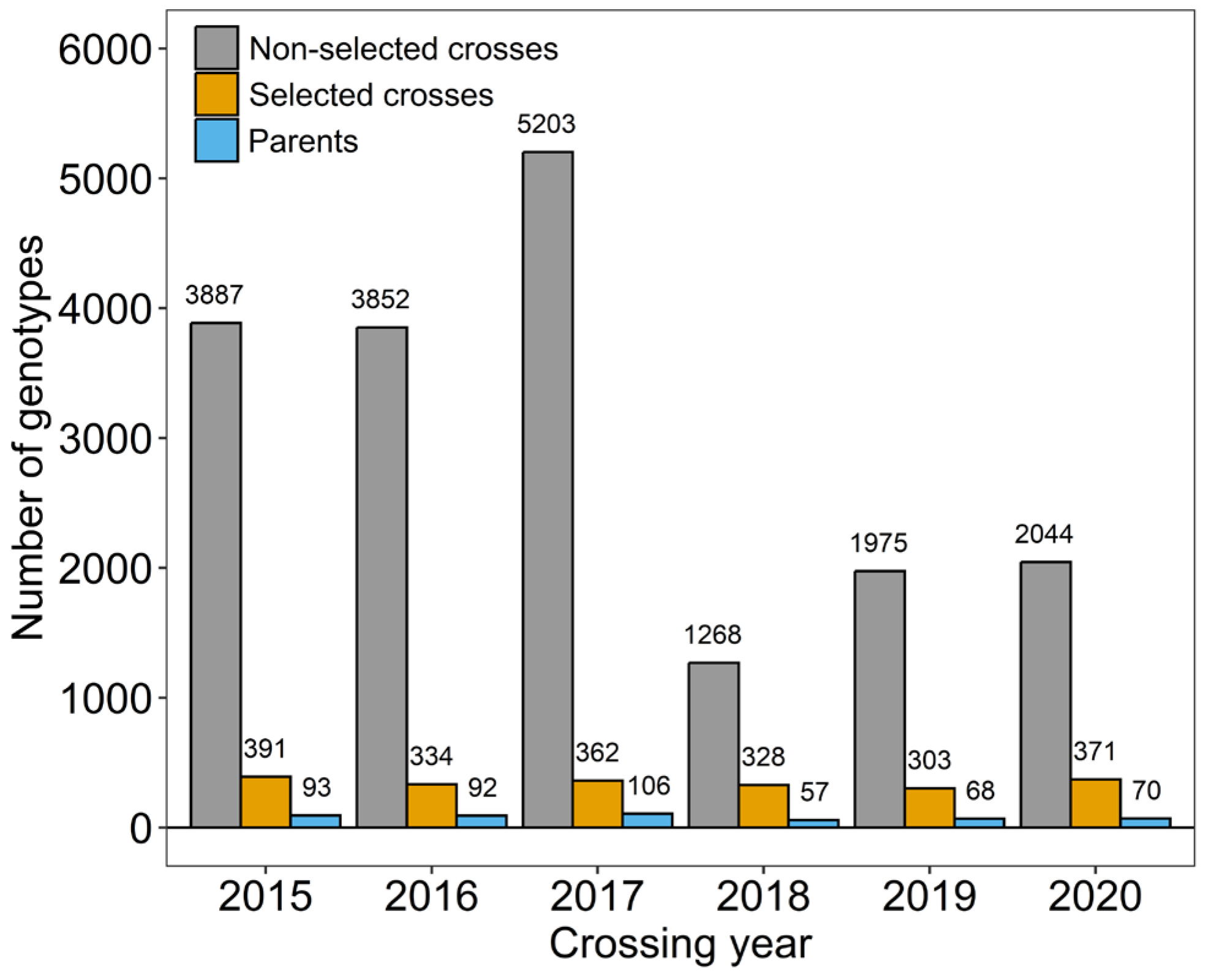
2.1. Plant Material, Classification of Breeder’s Decisions, and Genotypic Data
2.2. Forward Prediction of the Breeder’s Choice of Crosses
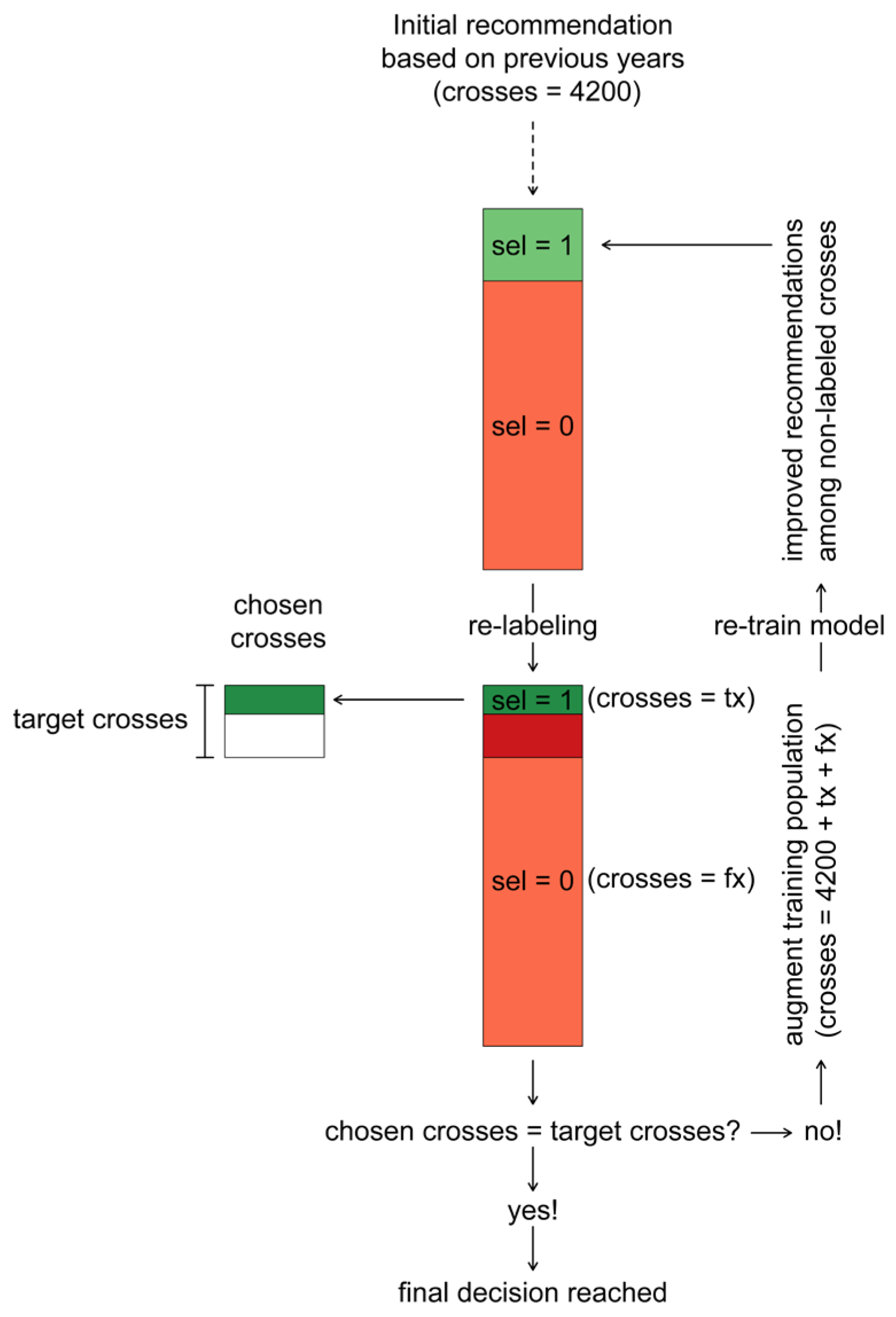
2.3. Integrating Model-Based Recommendations and Breeder’s Preferences
- In the first iteration the crosses with the highest probability of selection were recommended and labeled to fall into the class of selected crosses and all the other 1200 crosses to fall into the class of non-selected crosses .
- Based on the actual breeder’s choice among these recommended crosses, this labeling of was retained for a number of crosses corresponding to the true positives, while it was changed to for a number of crosses corresponding to the false positives.
- The true positive crosses were then added to the pool of chosen crosses .
- After (re)-labeling the crosses in the validation population in this way, they were used to augment the initial training population of crosses to a size of .
- Prediction models were then re-trained with this augmented training population to obtain recommendations for the remaining not yet labeled crosses.
- In the second iteration the crosses with the highest probability of selection were recommended and labeled to fall into the class of selected crosses , reducing the number of recommended crosses to the remaining difference towards the target of crosses.
- Steps 2–6 were repeated for iterations or until the target number of crosses was reached in the pool of chosen crosses
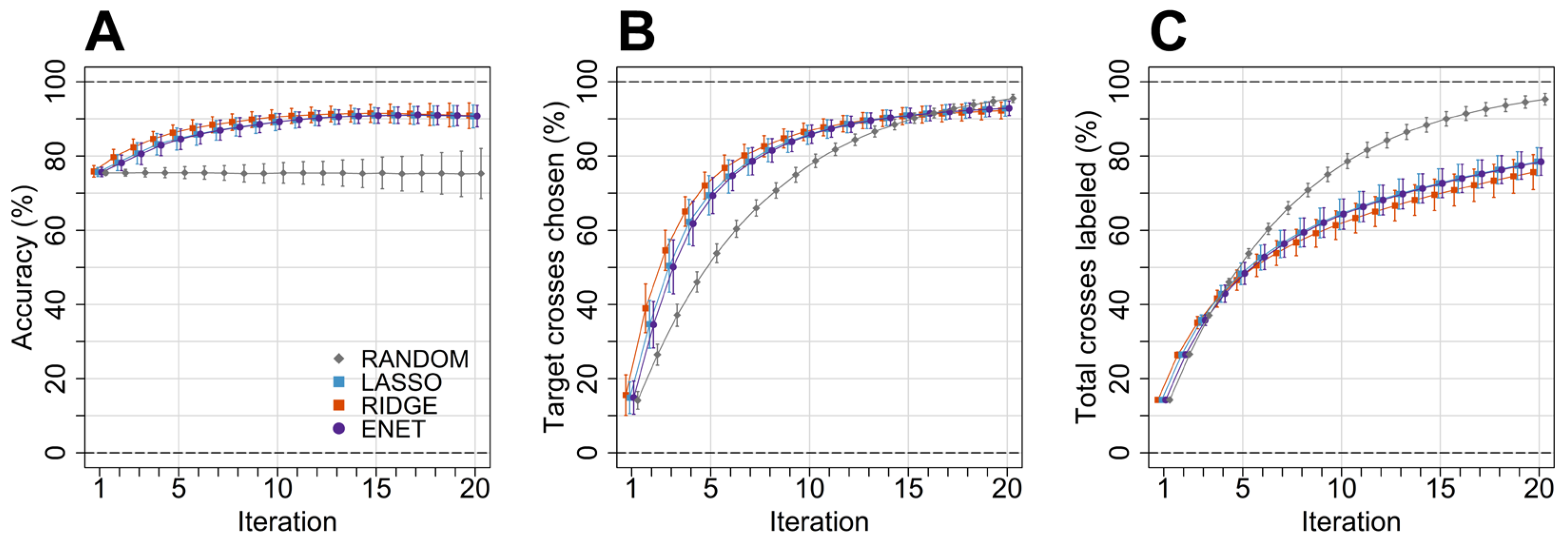
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Witcombe, J.R.; Gyawali, S.; Subedi, M.; Virk, D.S.; Joshi, K.D. Plant breeding can be made more efficient by having fewer, better crosses. BMC Plant Biol. 2013, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Witcombe, J.; Virk, D. Number of crosses and population size for participatory and classical plant breeding. Euphytica 2001, 122, 451–462. [Google Scholar] [CrossRef]
- van Ginkel, M.; Ortiz, R. Cross the Best with the Best, and Select the Best: HELP in Breeding Selfing Crops. Crop. Sci. 2018, 58, 17–30. [Google Scholar] [CrossRef]
- Busch, R.H.; Janke, J.C.; Frohberg, R.C. Evaluation of Crosses Among High and Low Yielding Parents of Spring Wheat (Triticum aestivum L.) and Bulk Prediction of Line Performance. Crop. Sci. 1974, 14, 47–50. [Google Scholar] [CrossRef]
- Miedaner, T.; Schneider, B.; Oettler, G. Means and variances for Fusarium head blight resistance of F2-derived bulks from winter triticale and winter wheat crosses. Euphytica 2006, 152, 405–411. [Google Scholar] [CrossRef]
- Utz, H.F.; Bohn, M.; Melchinger, A.E. Predicting progeny means and variances of winter wheat crosses from phenotypic values of their parents. Crop. Sci. 2001, 41, 1470–1478. [Google Scholar] [CrossRef]
- Déserts, A.D.D.; Durand, N.; Servin, B.; Goudemand-Dugué, E.; Alliot, J.-M.; Ruiz, D.; Charmet, G.; Elsen, J.-M.; Bouchet, S. Comparison of genomic-enabled cross selection criteria for the improvement of inbred line breeding populations. G3 Genes|Genomes|Genetics 2023, 13, jkad195. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Moreno-Amores, J.; Ametz, C.; Sparry, E.; Abel, E.; Ehn, M.; Bürstmayr, H. Balancing selection gain and genetic diversity in the genomic planning of crosses. Plant Breed. 2022, 141, 184–193. [Google Scholar] [CrossRef]
- Cowling, W.A. Sustainable plant breeding. Plant Breed. 2013, 132, 1–9. [Google Scholar] [CrossRef]
- De Beukelaer, H.; Badke, Y.; Fack, V.; De Meyer, G. Moving Beyond Managing Realized Genomic Relationship in Long-Term Genomic Selection. Genetics 2017, 206, 1127–1138. [Google Scholar] [CrossRef]
- Vanavermaete, D.; Fostier, J.; Maenhout, S.; De Baets, B. Adaptive scoping: Balancing short- and long-term genetic gain in plant breeding. Euphytica 2022, 218, 109. [Google Scholar] [CrossRef]
- Gorjanc, G.; Hickey, J.M. AlphaMate: A program for optimizing selection, maintenance of diversity and mate allocation in breeding programs. Bioinformatics 2018, 34, 3408–3411. [Google Scholar] [CrossRef]
- Jean, M.; Cober, E.; O'Donoughue, L.; Rajcan, I.; Belzile, F. Improvement of key agronomical traits in soybean through genomic prediction of superior crosses. Crop. Sci. 2021, 61, 3908–3918. [Google Scholar] [CrossRef]
- Lehermeier, C.; Teyssèdre, S.; Schön, C.-C. Genetic Gain Increases by Applying the Usefulness Criterion with Improved Variance Prediction in Selection of Crosses. Genetics 2017, 207, 1651–1661. [Google Scholar] [CrossRef]
- Miller, M.J.; Song, Q.; Fallen, B.; Li, Z. Genomic prediction of optimal cross combinations to accelerate genetic improvement of soybean (Glycine max). Front. Plant Sci. 2023, 14, 1171135. [Google Scholar] [CrossRef] [PubMed]
- Neyhart, J.L.; Smith, K.P. Validating Genomewide Predictions of Genetic Variance in a Contemporary Breeding Program. Crop. Sci. 2019, 59, 1062–1072. [Google Scholar] [CrossRef]
- Desta, Z.A.; Ortiz, R. Genomic selection: Genome-wide prediction in plant improvement. Trends Plant Sci. 2014, 19, 592–601. [Google Scholar] [CrossRef]
- Burke, R.; Felfernig, A.; Göker, M.H. Recommender Systems: An Overview. AI Mag. 2011, 32, 13–18. [Google Scholar] [CrossRef]
- Diversity Arrays Technology Pty Ltd. DArT P/L. 2020. Available online: https://www.diversityarrays.com/ (accessed on 4 February 2023).
- Stekhoven, D.J.; Bühlmann, P. Missforest-Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Z.; Liu, G.; Jiang, Y.; Maurer, H.P.; Würschum, T.; Matros, A.; Ebmeyer, E.; Schachschneider, R.; Kazman, E.; et al. Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. Proc. Natl. Acad. Sci. USA 2015, 112, 15624–15629. [Google Scholar] [CrossRef]
- Zhong, S.; Jannink, J.-L. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 2007, 177, 567–576. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 4 February 2023).
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Softw. 2023, 106, 1–31. [Google Scholar] [CrossRef]
- Rembe, M.; Zhao, Y.; Wendler, N.; Oldach, K.; Korzun, V.; Reif, J.C. The Potential of Genome-Wide Prediction to Support Parental Selection, Evaluated with Data from a Commercial Barley Breeding Program. Plants 2022, 11, 2564. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Chan, A.W.; Kulakow, P.; Rabbi, I.; Jannink, J.-L. Genomic mating in outbred species: Predicting cross usefulness with additive and total genetic covariance matrices. Genetics 2021, 219, iyab122. [Google Scholar] [CrossRef] [PubMed]
- Oget-Ebrad, C.; Heumez, E.; Duchalais, L.; Goudemand-Dugué, E.; Oury, F.-X.; Elsen, J.-M.; Bouchet, S. Validation of cross-progeny variance genomic prediction using simulations and experimental data in winter elite bread wheat. Theor. Appl. Genet. 2024, 137, 226. [Google Scholar] [CrossRef] [PubMed]
- Wartha, C.A.; Lorenz, A.J. Genomic predictions of genetic variances and correlations among traits for breeding crosses in soybean. Heredity 2024, 133, 173–185. [Google Scholar] [CrossRef]
- Pankaj, Y.K.; Kumar, R.; Gill, K.S.; Nagarajan, R. Unravelling QTLs for Non-Destructive and Yield-Related Traits Under Timely, Late and Very Late Sown Conditions in Wheat (Triticum aestivum L.). Plant Mol. Biol. Rep. 2024, 42, 369–382. [Google Scholar] [CrossRef]
- Borrenpohl, D.; Huang, M.; Olson, E.; Sneller, C. The value of early-stage phenotyping for wheat breeding in the age of genomic selection. Theor. Appl. Genet. 2020, 133, 2499–2520. [Google Scholar] [CrossRef]
- Neyhart, J.L.; Lorenz, A.J.; Smith, K.P. Multi-trait Improvement by Predicting Genetic Correlations in Breeding Crosses. G3 Genes Genomes Genet. 2019, 9, 3153–3165. [Google Scholar] [CrossRef]
- Raffo, M.A.; Sarup, P.; Guo, X.; Liu, H.; Andersen, J.R.; Orabi, J.; Jahoor, A.; Jensen, J. Improvement of genomic prediction in advanced wheat breeding lines by including additive-by-additive epistasis. Theor. Appl. Genet. 2022, 135, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Sneller, C.; Ignacio, C.; Ward, B.; Rutkoski, J.; Mohammadi, M. Using Genomic Selection to Leverage Resources among Breeding Programs: Consortium-Based Breeding. Agronomy 2021, 11, 1555. [Google Scholar] [CrossRef]
- Batmaz, Z.; Yurekli, A.; Bilge, A.; Kaleli, C. A review on deep learning for recommender systems: Challenges and remedies. Artif. Intell. Rev. 2019, 52, 1–37. [Google Scholar] [CrossRef]
- Roy, D.; Dutta, M. A systematic review and research perspective on recommender systems. J. Big Data 2022, 9, 1–36. [Google Scholar] [CrossRef]
- Günther, F.; Fritsch, S. neuralnet: Training of Neural Networks. R J. 2010, 2, 30–38. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Dodeja, L.; Tambwekar, P.; Hedlund-Botti, E.; Gombolay, M. Towards the design of user-centric strategy recommendation systems for collaborative Human–AI tasks. Int. J. Human-Computer Stud. 2024, 184, 103216. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S. Artificial Intelligence and Human Enhancement: Can AI Technologies Make Us More (Artificially) Intelligent? Camb. Q. Heal. Ethic 2024, 33, 76–88. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michel, S.; Löschenberger, F.; Ametz, C.; Bistrich, H.; Bürstmayr, H. Towards Streamlining the Choice of Crossing Combinations in Plant Breeding by Integrating Model-Based Recommendations and Plant Breeder’s Preferences. Crops 2025, 5, 5. https://doi.org/10.3390/crops5010005
Michel S, Löschenberger F, Ametz C, Bistrich H, Bürstmayr H. Towards Streamlining the Choice of Crossing Combinations in Plant Breeding by Integrating Model-Based Recommendations and Plant Breeder’s Preferences. Crops. 2025; 5(1):5. https://doi.org/10.3390/crops5010005
Chicago/Turabian StyleMichel, Sebastian, Franziska Löschenberger, Christian Ametz, Herbert Bistrich, and Hermann Bürstmayr. 2025. "Towards Streamlining the Choice of Crossing Combinations in Plant Breeding by Integrating Model-Based Recommendations and Plant Breeder’s Preferences" Crops 5, no. 1: 5. https://doi.org/10.3390/crops5010005
APA StyleMichel, S., Löschenberger, F., Ametz, C., Bistrich, H., & Bürstmayr, H. (2025). Towards Streamlining the Choice of Crossing Combinations in Plant Breeding by Integrating Model-Based Recommendations and Plant Breeder’s Preferences. Crops, 5(1), 5. https://doi.org/10.3390/crops5010005





