Whole Genome Sequence-Based Classification of Nonomuraea marmarensis sp. nov., Isolated from Island Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Cultivation
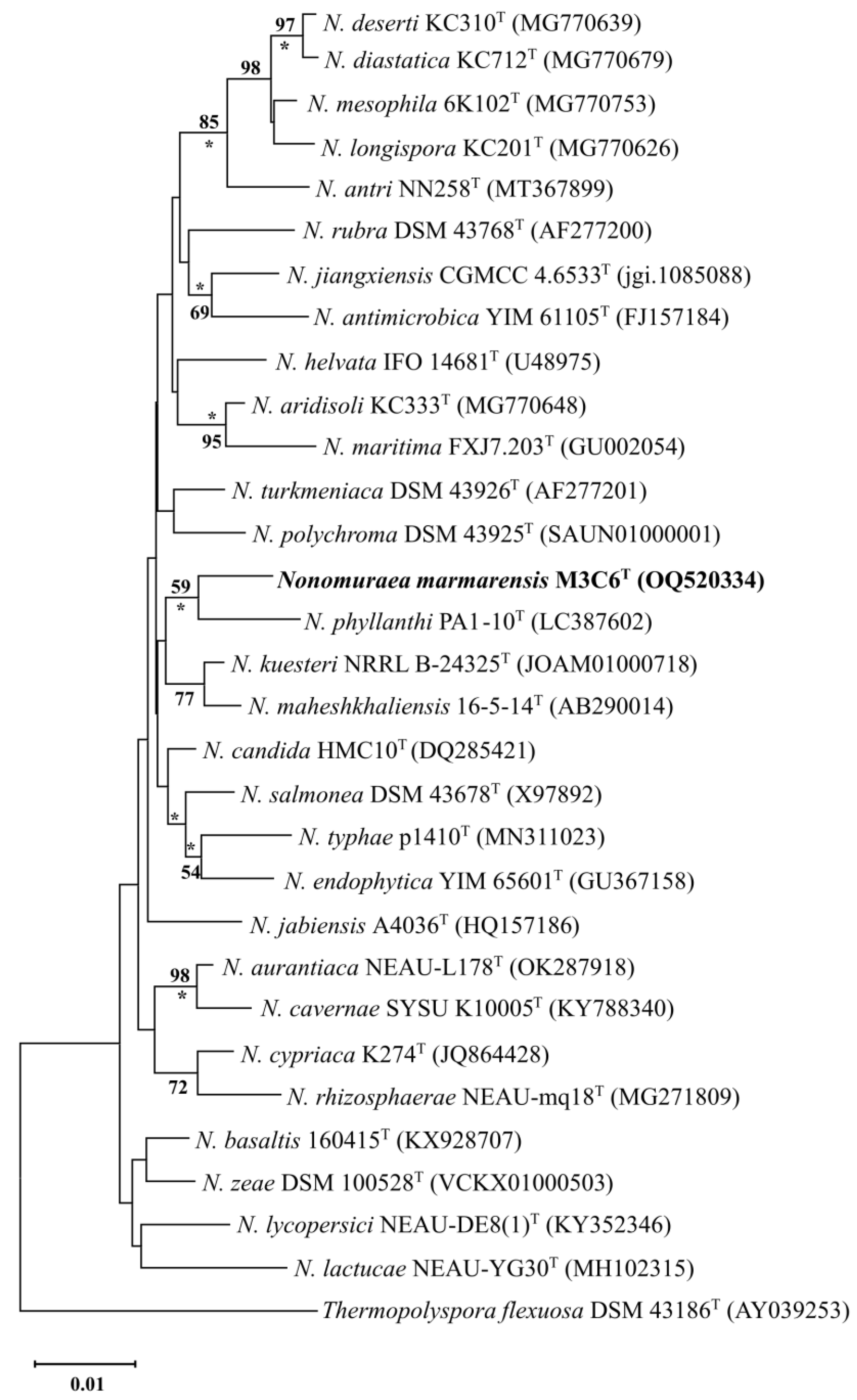
2.2. 16S rRNA Gene Phylogeny
2.3. Phylogeny and Genome Features Based on Whole-Genome Sequencing
2.4. Morphological, Cultural, Physiological and Biochemical Characteristics
2.5. Chemotaxonomy
3. Results and Discussion
3.1. Genotypic and Phylogenetic Analysis

3.2. Morphology, Physiology and Biochemical Analysis
3.3. Chemotaxonomic Characterization
4. Conclusions
Description of Nonomuraea marmarensis sp. nov.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Wang, Y.; Ruan, J. Reclassification of Thermomonospora and Microtetraspora. Int. J. Syst. Bacteriol. 1998, 48, 411–422. [Google Scholar] [CrossRef]
- Chiba, S.; Suzuki, M.; Ando, K. Taxonomic re-evaluation of ‘Nocardiopsis’ sp. K-252T (= NRRL 15532T): A proposal to transfer this strain to the genus Nonomuraea as Nonomuraea longicatena sp. nov. Int. J. Syst. Bacteriol. 1999, 49, 1623–1630. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nonomura, H.; Ohara, Y. Distribution of actinomycetes in soil. XI. Some new species of the genus Actinomadura Lechevalier et al. J. Ferment. Technol. 1971, 49, 904–912. [Google Scholar]
- Lechevalier, M.P.; Lechevalier, H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 1970, 20, 435–443. [Google Scholar] [CrossRef]
- Lechevalier, M.P.; De Bievre, C.; Lechevalier, H. Chemotaxonomy of aerobic actinomycetes: Phospholipid composition. Biochem. Syst. Ecol. 1977, 5, 249–260. [Google Scholar] [CrossRef]
- Kroppenstedt, R.M. Fatty acid and menaquinone analysis of actinomycetes and related organisms. In Chemical Methods in Bacterial Systematics; Goodfellow, M., Minnikin, D.E., Eds.; Academic Press: London, UK, 1985; pp. 173–199. [Google Scholar]
- Kämpfer, P. Genus VI. Nonomuraea. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Whitman, W.B., Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Ludwig, W., Suzuki, K.-I., Parte, A., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 1844–1861. [Google Scholar]
- Willdigg, J.R.; Helmann, J.D. Mini review: Bacterial membrane composition and its modulation in response to stress. Front. Mol. Biosci. 2021, 8, 634438. [Google Scholar] [CrossRef]
- Saricaoglu, S.; Nouioui, I.; Ay, H.; Saygin, H.; Inan Bektas, K.; Guven, K.; Cetin, D.; Klenk, H.P.; Isik, K.; Sahin, N. Nonomuraea insulae sp. nov., isolated from forest soil. Antonie Van Leeuwenhoek 2018, 111, 2051–2059. [Google Scholar] [CrossRef]
- Veyisoglu, A. Nonomuraea cypriaca sp. nov., isolated from soil. Arch. Microbiol. 2021, 203, 2639–2645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.R.; Zhang, Y.J.; Zhao, J.W.; Liu, C.X.; Wang, S.R.; Yang, L.Y.; He, H.R.; Xiang, W.; Wang, X.S.; Wang, X.J. Nonomuraea fuscirosea sp. nov., an actinomycete isolated from the rhizosphere soil of rehmannia (Rehmannia glutinosa Libosch). Int. J. Syst. Evol. Microbiol. 2014, 64, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Qin, X.; Tian, X.; Zhao, Y.; Yang, T.; Huang, J. Inoculating with the microbial agents to start up the aerobic composting of mushroom residue and wood chips at low temperature. J. Environ. Chem. Eng. 2021, 9, 105294. [Google Scholar] [CrossRef]
- Casciello, C.; Tonin, F.; Berini, F.; Fasoli, E.; Marinelli, F.; Pollegioni, L.; Rosini, E. A valuable peroxidase activity from the novel species Nonomuraea gerenzanensis growing on alkali lignin. Biotechnol. Rep. 2017, 13, 49–57. [Google Scholar] [CrossRef]
- Kontro, M.H.; Yaradoddi, J.S.; Banapurmath, N.R.; Ganachari, S.V.; Hungund, B.S. Biotechnological importance of Actinomycetes. In Actinobacteria: Ecology, Diversity, Classification and Extensive Applications; Yaradoddi, J.S., Kontro, M.H., Ganachari, S.V., Eds.; Springer Nature: Singapore, 2022; pp. 271–290. [Google Scholar]
- Zhang, S.; Wang, L.; Zhou, B.; Zhang, D.; Tang, G.; Guo, L. Characteristics of humification, functional enzymes and bacterial community metabolism during manganese dioxide-added composting of municipal sludge. Environ. Res. 2024, 252, 119151. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhu, A.; Hou, J.; Wang, L.; Zhang, R.; Li, J.; Guo, Y.; Chu, Y. Nonomuraea sediminis sp. nov., a novel actinobacterium with antimicrobial activity, isolated from sediment of Dianchi Lake. Arch. Microbiol. 2023, 205, 91. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.Z.; Hua, Z.S.; Han, M.X.; Zhang, Z.T.; Wang, Y.H.; Yang, Z.W.; Zhang, W.Q.; Li, M.X.; Li, W.-J. Nonomuraea cavernae sp. nov., a novel actinobacterium. Int. J. Syst. Evol. Microbiol. 2017, 67, 2879–2884. [Google Scholar] [CrossRef]
- Niemhom, N.; Chutrakul, C.; Suriyachadkun, C.; Thawai, C. Nonomuraea stahlianthi sp. nov., an endophytic actinomycete isolated from the stem of Stahlianthus campanulatus. Int. J. Syst. Evol. Microbiol. 2017, 67, 2879–2884. [Google Scholar] [CrossRef]
- Rachniyom, H.; Matsumoto, A.; Indananda, C.; Duangmal, K.; Takahashi, Y.; Thamchaipenet, A. Nonomuraea syzygii sp. nov., an endophytic actinomycete isolated from the roots of a jambolan plum tree (Syzygium cumini L. Skeels). Int. J. Syst. Evol. Microbiol. 2015, 65, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Sungthong, R.; Nakaew, N. The genus Nonomuraea: A review of a rare actinomycete taxon for novel metabolites. J. Basic Microbiol. 2015, 55, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yamada, K.; Zhou, T.; Harunari, E.; Igarashi, Y.; Terahara, T.; Imada, C. Akazamicin, a cytotoxic aromatic polyketide from marine-derived Nonomuraea sp. J. Antibiot. 2019, 72, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Duangupama, T.; Pittayakhajonwut, P.; Intaraudom, C.; Suriyachadkun, C.; Tadtong, S.; Kuncharoen, N.; Thawai, C. Pradimicin U, a promising antimicrobial agent isolated from a newly found Nonomuraea composti sp. nov. Sci. Rep. 2023, 13, 10942. [Google Scholar] [CrossRef] [PubMed]
- Saygin, H.; Nouioui, I.; Ay, H.; Guven, K.; Cetin, D.; Klenk, H.P.; Goodfellow, M.; Sahin, N. Polyphasic classification of Nonomuraea strains isolated from the Karakum Desert and description of Nonomuraea deserti sp. nov., Nonomuraea diastatica sp. nov., Nonomuraea longispora sp. nov. and Nonomuraea mesophila sp. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 636–647. [Google Scholar] [CrossRef]
- Yushchuk, O.; Vior, N.M.; Andreo-Vidal, A.; Berini, F.; Rückert, C.; Busche, T.; Kalinowski, J.; Sosio, M.; Luzhetskyy, A.; Marinelli, F. Genomic-led discovery of a novel glycopeptide antibiotic by Nonomuraea coxensis DSM 45129. ACS Chem. Biol. 2021, 16, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Ustaömer, P.A.; Ustaömer, T.; Collins, A.S.; Reischpeitsch, J. Lutetian Arc-Type Magmatism along the Southern Eurasian Margin: New U-Pb LA-ICPMS and Whole-Rock Geochemical Data from Marmara Island, NW Turkey. Miner. Pet. 2009, 96, 177–196. [Google Scholar] [CrossRef]
- Bulut, G. Medicinal and wild food plants of Marmara Island (Balikesir-Turkey). Acta Soc. Bot. Pol. 2016, 85, 2. [Google Scholar] [CrossRef][Green Version]
- Tuzlacı, E. The flora of Marmara Island. J. Fac. Pharm. Istanbul Univ. 1981, 17, 138–154. [Google Scholar]
- Tuzlacı, E. Observations and New Floristic Findings on the Vegetation of Marmara Island. In Proceedings of the TÜBİTAK VII. Science Congress Mathematics, Physics, and Biological Sciences Research Group Presentations, Ankara, Turkey, 6–10 October 1980; pp. 749–758. [Google Scholar]
- Weyland, H. Actinomycetes in North Sea and Atlantic Ocean sediments. Nature 1969, 223, 858. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Chun, J.; Goodfellow, M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 1995, 45, 240–245. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogeny: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L. Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J. Bacteriol. 1949, 57, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.A. The Actinomycetes: A Summary of Current Knowledge; Ronald Press: New York, NY, USA, 1967. [Google Scholar]
- Waksman, S.A. The Actinomycetes, Vol. II: Classification, Identification and Descriptions of Genera and Species; Williams & Wilkins: Baltimore, MD, USA, 1961. [Google Scholar]
- Kelly, K.L. Inter-Society Color Council-National Bureau of Standards Color-Name Charts Illustrated with Centroid Colors; U.S. Government Printing Office: Washington, DC, USA, 1964.
- Williams, S.T.; Goodfellow, M.; Alderson, G.; Wellington, E.M.H.; Sneath, P.H.A.; Sackin, M.J. Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 1983, 129, 1743–1813. [Google Scholar] [CrossRef]
- Collins, C.H.; Lyne, P.M.; Grange, J.M. Microbiological Methods, 8th ed.; Arnold, a member of the Hodder Headline Group: London, UK, 2004. [Google Scholar]
- Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. Technical Note 101, MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Kämpfer, P.; Kroppenstedt, R.M. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 1996, 42, 989–1005. [Google Scholar] [CrossRef]
- Collins, M.D.; Pirouz, T.; Goodfellow, M.; Minnikin, D.E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 1977, 100, 221–230. [Google Scholar] [CrossRef]
- Kroppenstedt, R.M. Separation of bacterial menaquinones by HPLC using reverse phase (RP-18) and a silver loaded ion exchanger. J. Liquid Chromatogr. 1982, 5, 2359–2387. [Google Scholar] [CrossRef]
- Staneck, J.L.; Roberts, G.D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 1974, 28, 226–231. [Google Scholar] [CrossRef]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, K.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Kroppenstedt, R.M.; Goodfellow, M. The family Thermomonosporaceae: Actinocorallia, Actinomadura, Spirillispora and Thermomonospora. In The Prokaryotes Archaea and Bacteria: Firmicutes, Actinomycetes, 3rd ed.; Dworkin, M., Falkow, S., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 3, pp. 682–724. [Google Scholar]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Drautz, H.; Keller-Schierlein, W.; Zähner, H. Metabolic products of microorganisms, 149. Lysolipin I, a new antibiotic from Streptomyces violaceoniger (author’s transl). Arch. Microbiol. 1975, 106, 175–190. [Google Scholar] [CrossRef]
- Szczurek, A.; Maciejewska, M.; Kabsch-Korbutowicz, M.; Wolska, M.; Solipiwko-Pieścik, A. Geosmin and 2-Methylisoborneol Detection in Water Using Semiconductor Gas Sensors. Electronics 2023, 13, 63. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Nguyen, C.T.; Choi, Y.S.; Dhakal, D.; Kim, T.S.; Jung, H.J.; Yamaguchi, T.; Sohng, J.K. Identification and enhancing production of a novel macrolide compound in engineered Streptomyces peucetius. RSC Adv. 2021, 11, 3168–3173. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Klykleung, N.; Yuki, M.; Kudo, T.; Ohkuma, M.; Phongsopitanun, W.; Phongsopitanun, A.; Duangmal, K. Nonomuraea phyllanthi sp. nov., an endophytic actinomycete isolated from the leaf of Phyllanthus amarus. Arch. Microbiol. 2020, 202, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Saricaoglu, S.; Inan, B.; Guven, K.; Saygin, H.; Isik, K.; Cetin, D.; Bektas, K.I.; Sahin, N. Nonomuraea basaltis sp. nov., a siderophore-producing actinobacteria isolated from surface soil of basaltic parent material. Arch. Microbiol. 2020, 202, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.; Ding, Y.; Gao, Y.; Ruan, J.; Huang, Y. Nonomuraea jiangxiensis sp. nov., isolated from acidic soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 1409–1413. [Google Scholar] [CrossRef] [PubMed]

| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| GenBank number | JBICRM000000000 | VCJS00000000 | VCKY00000000 | VDLX00000000 | FNDJ00000000 |
| Genome size (bp) | 10,380,745 | 14,334,648 | 11,350,886 | 11,027,144 | 11,808,550 |
| Number of contigs | 142 | 671 | 592 | 64 | 68 |
| Genome coverage | 90× | 30× | 30× | 300× | 67× |
| G+C content (%) | 69.5 | 69.6 | 69.7 | 71.2 | 70.6 |
| Genes (RNA) | 87 | 81 | 75 | 78 | 87 |
| Genes (total) | 10,066 | 13,994 | 10,944 | 10,232 | 10,925 |
| Genes (coding) | 9827 | 13,067 | 10,275 | 9727 | 10,711 |
| Longest contig size | 618,571 | 176,753 | 143,656 | 1,156,183 | 759,783 |
| N50 value | 213,670 | 50,765 | 38,021 | 409,871 | 367,974 |
| L50 value | 15 | 86 | 96 | 9 | 12 |
| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| NaCI tolerance (%, w/v) | 0–1 | 0–2 | 0–1 | 0–4 | 0–5 |
| pH tolerance | 5–11 | 6–9 | 7–9 | 5–9 | 6–10 |
| Temperature range (°C) | 28–37 | 10–40 | 20–40 | 20–40 | 28–37 |
| Allantoin hydrolysis | − | + | − | − | − |
| Urea hydrolysis | + | − | + | + | − |
| Nitrate reduction | − | + | − | + | + |
| Nitrogen source utilization (0.1%, w/v) | |||||
| L-Arginine | + | − | + | + | + |
| Glycine | + | − | + | + | + |
| L-Histidine | + | − | − | + | − |
| L-Proline | − | + | + | + | + |
| L-Threonine | − | + | + | − | + |
| Carbon source utilization (1.0%, w/v) | |||||
| D-Arabinose | + | + | − | − | − |
| D-Cellobiose | + | + | + | − | + |
| D-Fructose | + | − | + | − | + |
| D-Mannose | + | + | − | + | − |
| D-Melibiose | − | + | + | + | − |
| L-Sorbose | − | − | + | + | − |
| L-Rhamnose | + | + | − | − | − |
| Xylitol | − | − | + | − | + |
| Degradation of (%, w/v) | |||||
| Guanine (0.05%) | + | + | − | − | − |
| Hypoxanthine (0.4%) | − | + | − | + | + |
| Tween 20 (1.0%) | − | − | + | − | − |
| Tween 40 (1.0%) | − | − | + | + | − |
| Tween 80 (1.0%) | − | − | + | − | − |
| Xanthine (0.4%) | − | − | + | − | − |
| Major menaquinones (>10%) | MK-9(H4) | MK-9(H4), MK-9(H6) * | ND | MK-9(H4), MK-9(H2) # | MK-9(H4), MK-9(H6) γ |
| Major polar lipids | DPG, PE, OH-PE, 2OH-PE, PG, PI, PME | DPG, PG * | ND | DPG, PE, lyso-PE, PG, PIM, PME# | DPG, PE, PG, PI, PME, OH-PME γ |
| Major fatty acids (>10%) | iso C16:0, iso C16:0 2OH | iso C16:0, C17:0 10-methyl, iso C16:1 G | iso C16:0, C17:0 10-methyl, iso C16:0 2OH | iso C15:0, iso C16:0, C17:0 10-methyl | iso C16:0, iso C16:1 G, C17:1 ω6c γ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topkara, A.R.; Saygin, H.; Saricaoglu, S.; Veyisoglu, A.; Tokatli, A.; Guven, K.; Cetin, D.; Isik, K. Whole Genome Sequence-Based Classification of Nonomuraea marmarensis sp. nov., Isolated from Island Soil. Taxonomy 2025, 5, 5. https://doi.org/10.3390/taxonomy5010005
Topkara AR, Saygin H, Saricaoglu S, Veyisoglu A, Tokatli A, Guven K, Cetin D, Isik K. Whole Genome Sequence-Based Classification of Nonomuraea marmarensis sp. nov., Isolated from Island Soil. Taxonomy. 2025; 5(1):5. https://doi.org/10.3390/taxonomy5010005
Chicago/Turabian StyleTopkara, Ahmet Ridvan, Hayrettin Saygin, Salih Saricaoglu, Aysel Veyisoglu, Ali Tokatli, Kiymet Guven, Demet Cetin, and Kamil Isik. 2025. "Whole Genome Sequence-Based Classification of Nonomuraea marmarensis sp. nov., Isolated from Island Soil" Taxonomy 5, no. 1: 5. https://doi.org/10.3390/taxonomy5010005
APA StyleTopkara, A. R., Saygin, H., Saricaoglu, S., Veyisoglu, A., Tokatli, A., Guven, K., Cetin, D., & Isik, K. (2025). Whole Genome Sequence-Based Classification of Nonomuraea marmarensis sp. nov., Isolated from Island Soil. Taxonomy, 5(1), 5. https://doi.org/10.3390/taxonomy5010005






