Corundum Particles as Trypsin Carrier for Efficient Protein Digestion
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
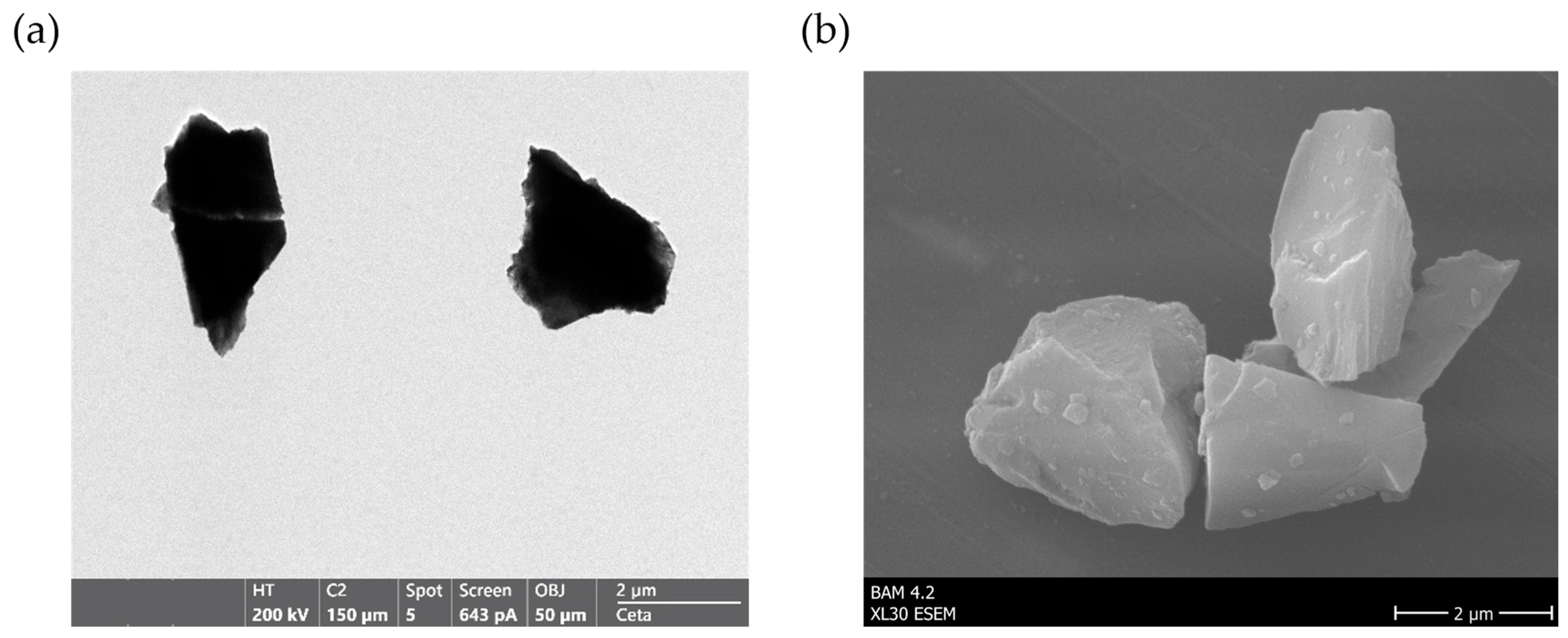
2.2. Particle Characterization via TEM, ESEM, DLS and Particle Size Distribution
2.3. Recombinant Production of Porcine Trypsin
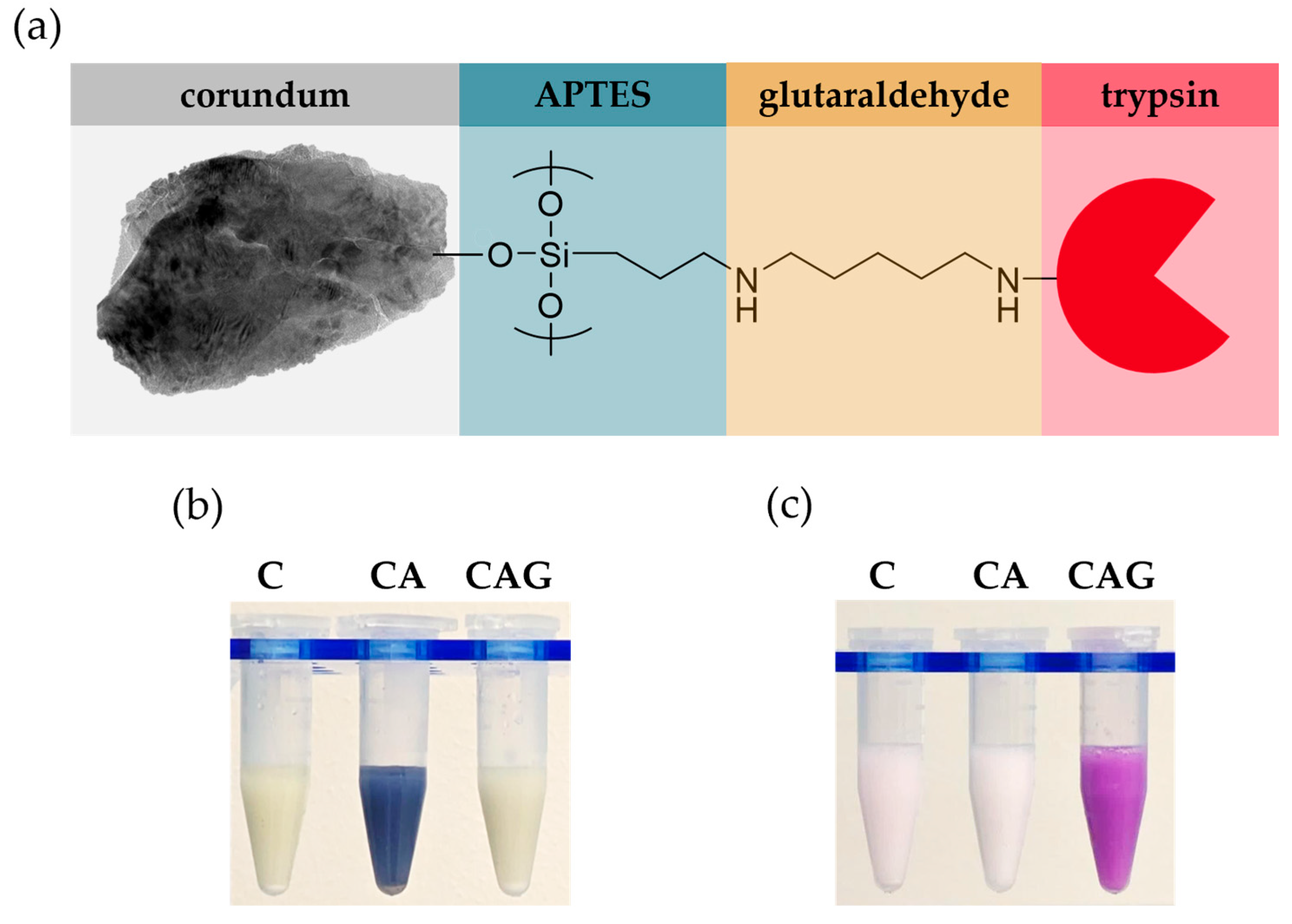
2.4. Surface Modification and Trypsin-Immobilization of Corundum
2.5. Colorimetric Verification of Surface Functionalization
2.5.1. Kaiser Test
2.5.2. Schiff Test
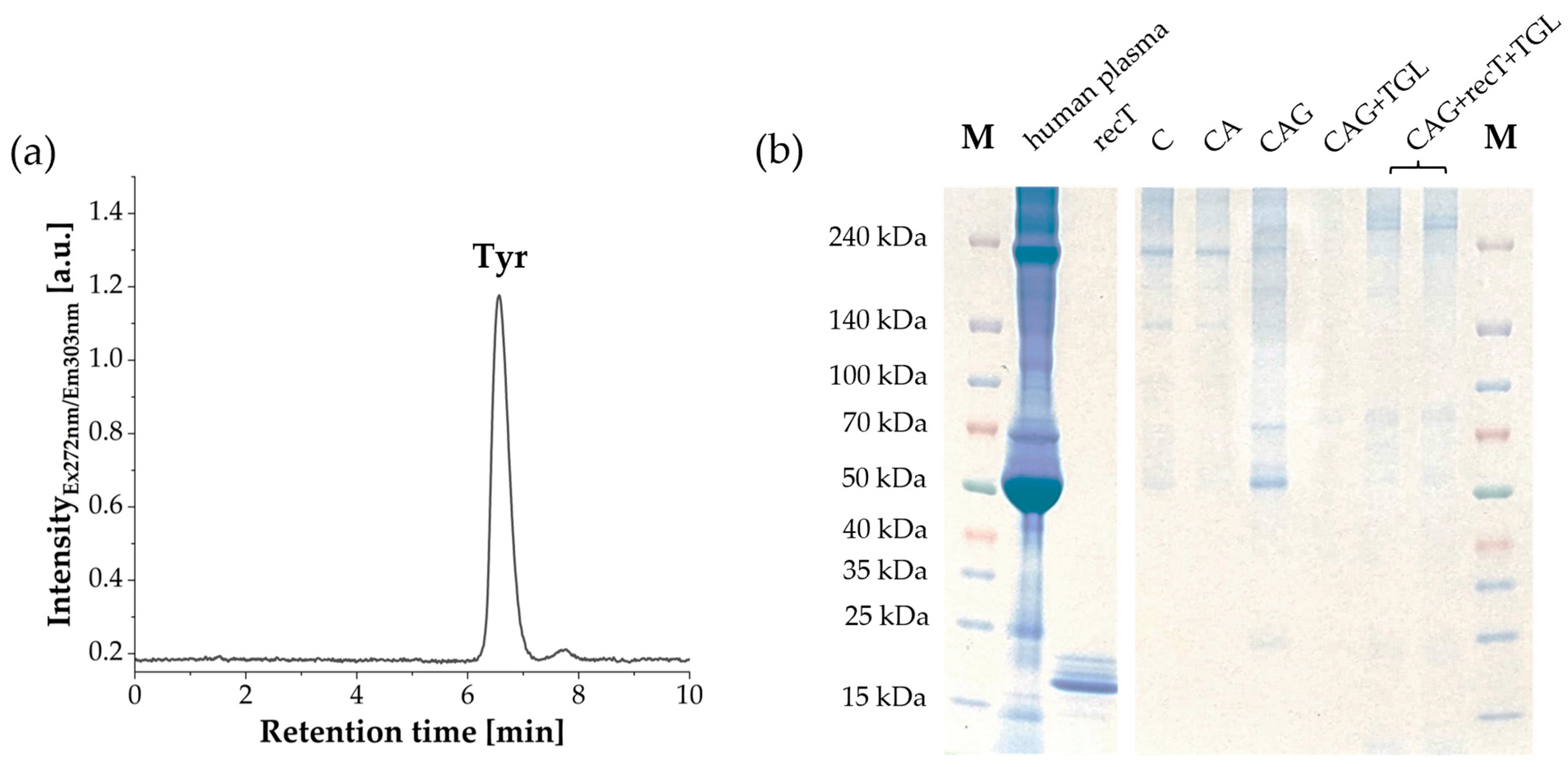
2.6. Aromatic Amino Acid Analysis (AAAA)
2.7. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
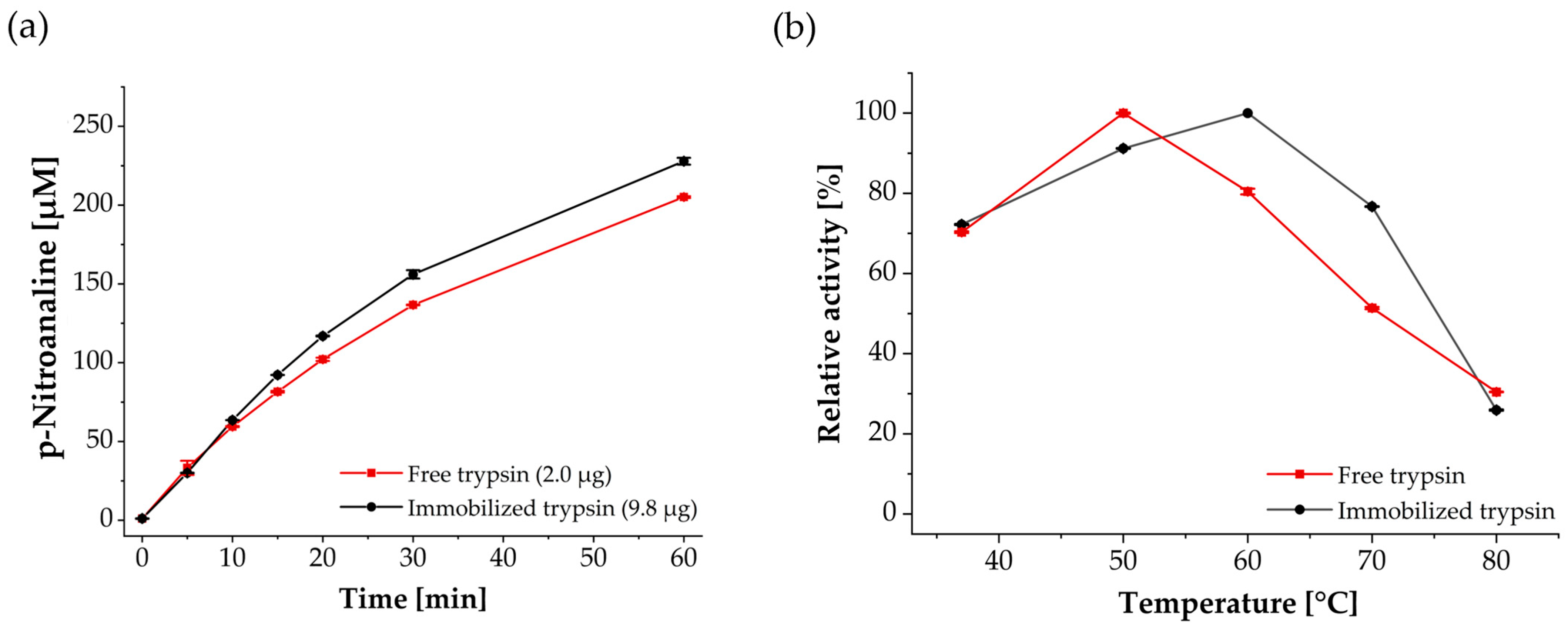
2.8. Trypsin Activity Assay Using Benzoyl-DL-arginine-p-nitroanilide (BAPNA)
2.9. Antibody Digestion and LC-MS/MS-Based Quantitative NISTmAb Analysis
2.10. MALDI-TOF MS-Based Antibody Fingerprinting and Identification of Herceptin
3. Results
3.1. Characterization of Raw and Surface-Modified Corundum Particles
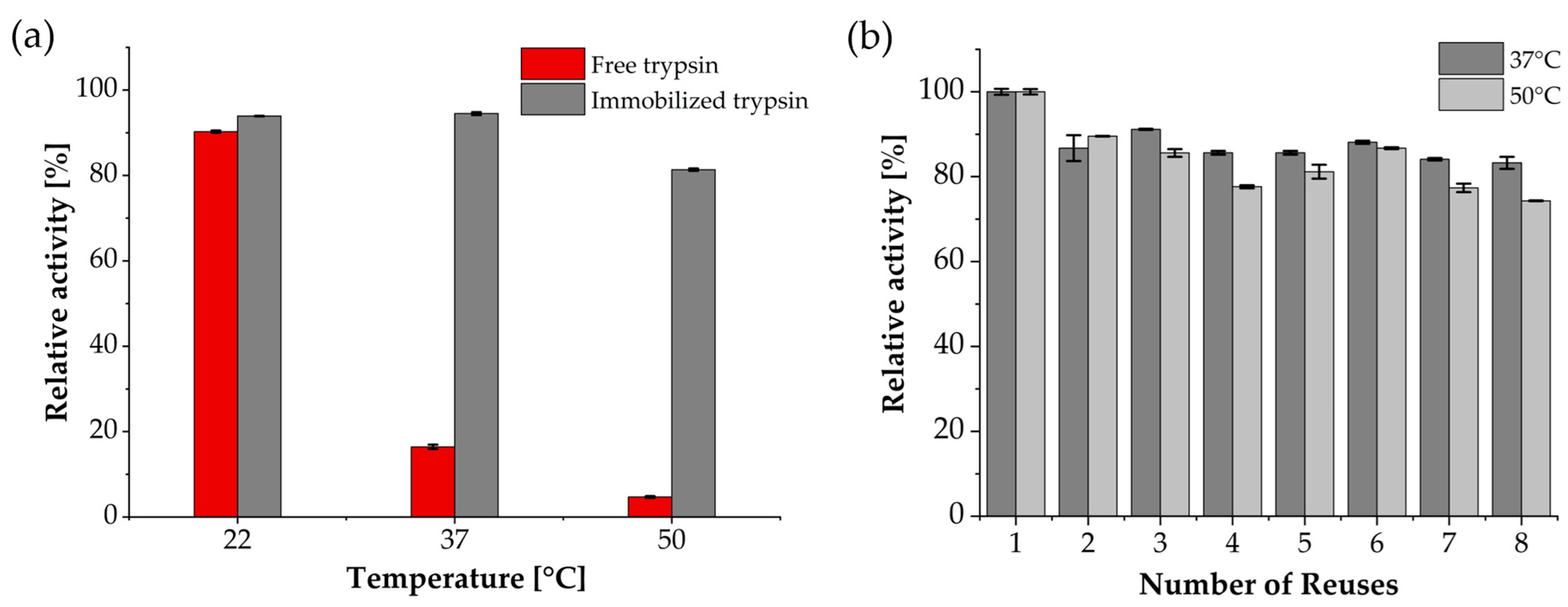
3.2. Enzymatic, Temperature-Dependent Activity and Reusability
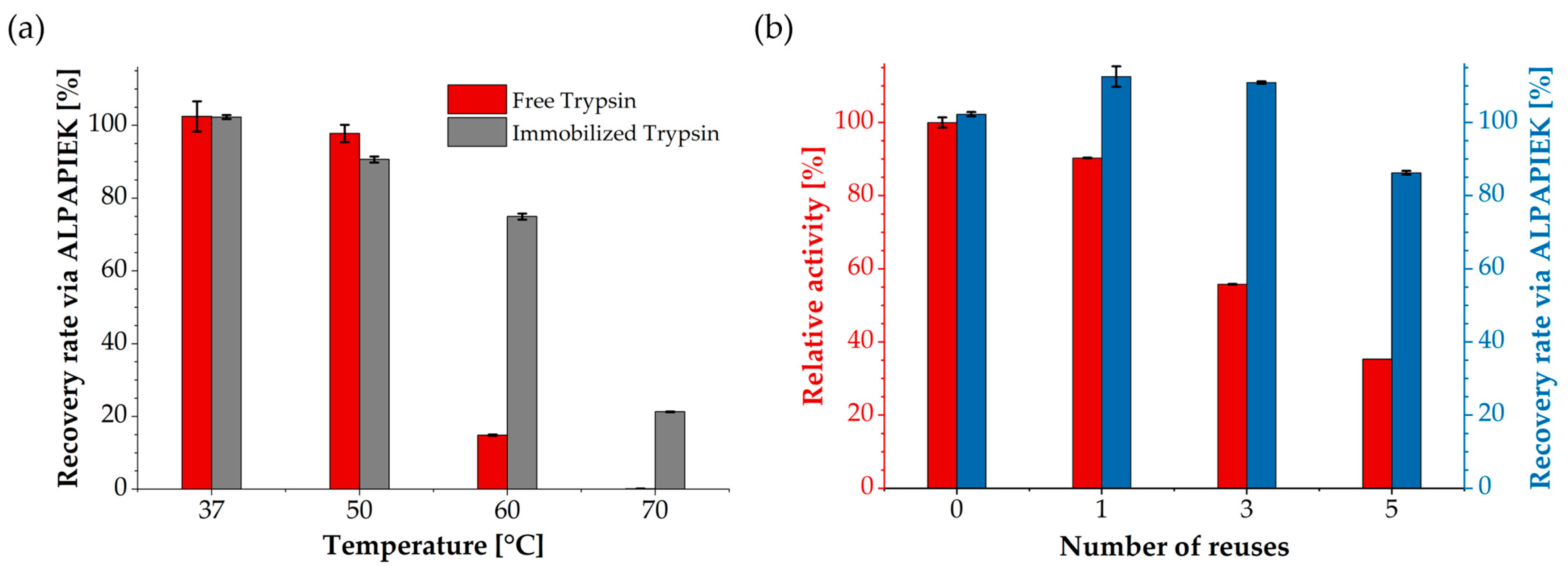
3.3. Application for LC-MS/MS-Based Quantification of NISTmAb
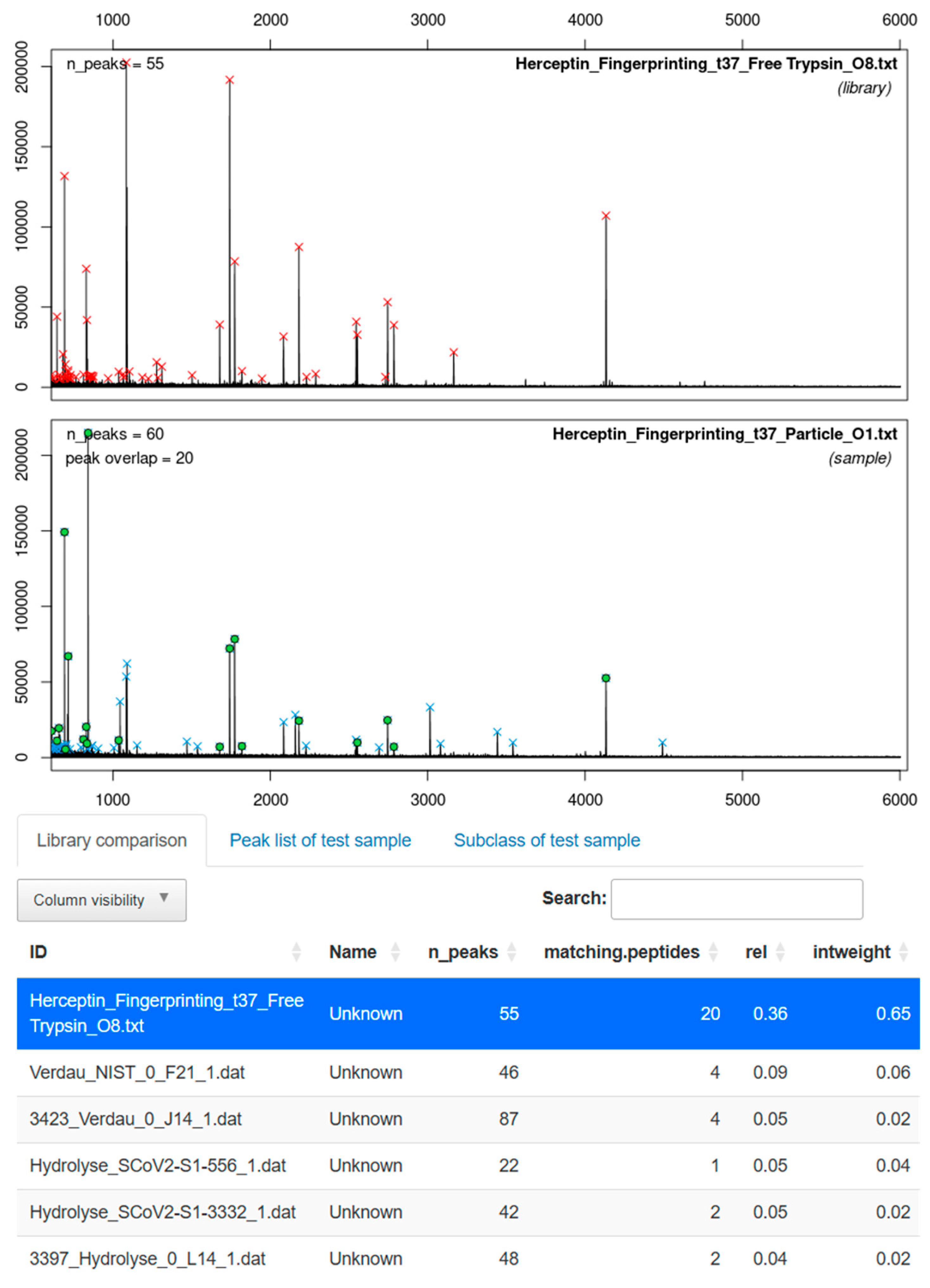
3.4. Application for MALDI-TOF MS-Based Identification of Herceptin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cavalcante, F.T.; Cavalcante, A.L.; de Sousa, I.G.; Neto, F.S.; dos Santos, J.C. Current status and future perspectives of supports and protocols for enzyme immobilization. Catalysts 2021, 11, 1222. [Google Scholar] [CrossRef]
- Bié, J.; Sepodes, B.; Fernandes, P.C.; Ribeiro, M.H. Enzyme immobilization and co-immobilization: Main framework, advances and some applications. Processes 2022, 10, 494. [Google Scholar] [CrossRef]
- Guisan, J.M.; Fernandez-Lorente, G.; Rocha-Martin, J.; Moreno-Gamero, D. Enzyme immobilization strategies for the design of robust and efficient biocatalysts. Curr. Opin. Green Sustain. Chem. 2022, 35, 100593. [Google Scholar] [CrossRef]
- Maghraby, Y.R.; El-Shabasy, R.M.; Ibrahim, A.H.; Azzazy, H.M.E.-S. Enzyme immobilization technologies and industrial applications. ACS Omega 2023, 8, 5184–5196. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Budžaki, S.; Miljić, G.; Sundaram, S.; Tišma, M.; Hessel, V. Cost analysis of enzymatic biodiesel production in small-scaled packed-bed reactors. Appl. Energy 2018, 210, 268–278. [Google Scholar] [CrossRef]
- Federsel, H.-J.; Moody, T.S.; Taylor, S.J. Recent trends in enzyme immobilization—Concepts for expanding the biocatalysis toolbox. Molecules 2021, 26, 2822. [Google Scholar] [CrossRef]
- Asanomi, Y.; Yamaguchi, H.; Miyazaki, M.; Maeda, H. Enzyme-immobilized microfluidic process reactors. Molecules 2011, 16, 6041–6059. [Google Scholar] [CrossRef]
- DiCosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar] [CrossRef]
- Liese, A.; Hilterhaus, L. Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [Google Scholar] [CrossRef]
- Lee, H.; Hong, Y.J.; Baik, S.; Hyeon, T.; Kim, D.H. Enzyme-based glucose sensor: From invasive to wearable device. Adv. Healthc. Mater. 2018, 7, 1701150. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Lee, S.H.; Lee, U.J.; Fermin, C.D.; Kim, M. Immobilized enzymes in biosensor applications. Materials 2019, 12, 121. [Google Scholar] [CrossRef]
- Naldi, M.; Tramarin, A.; Bartolini, M. Immobilized enzyme-based analytical tools in the-omics era: Recent advances. J. Pharm. Biomed. Anal. 2018, 160, 222–237. [Google Scholar] [CrossRef]
- Nagy, C.; Szabo, R.; Gaspar, A. Microfluidic immobilized enzymatic reactors for proteomic analyses—Recent developments and trends (2017–2021). Micromachines 2022, 13, 311. [Google Scholar] [CrossRef]
- Yao, Z.; Li, Y.; Xu, W. Micro-immobilized enzyme reactors for mass spectrometry proteomics. Analyst 2025, 150, 3000–3010. [Google Scholar] [CrossRef] [PubMed]
- Basso, A.; Froment, L.; Hesseler, M.; Serban, S. New highly robust divinyl benzene/acrylate polymer for immobilization of lipase CALB. Eur. J. Lipid Sci. Technol. 2013, 115, 468–472. [Google Scholar] [CrossRef]
- dos Santos, J.C.; Rueda, N.; Barbosa, O.; del Carmen Millán-Linares, M.; Pedroche, J.; del Mar Yuste, M.; Goncalves, L.R.; Fernandez-Lafuente, R. Bovine trypsin immobilization on agarose activated with divinylsulfone: Improved activity and stability via multipoint covalent attachment. J. Mol. Catal. B Enzym. 2015, 117, 38–44. [Google Scholar] [CrossRef]
- Lavlinskaya, M.S.; Sorokin, A.V.; Holyavka, M.G.; Zuev, Y.F.; Artyukhov, V.G. Cellulose and cellulose-based materials for enzyme immobilization: A review. Biophys. Rev. 2025, 17, 1–37. [Google Scholar] [CrossRef]
- Hartmann, M.; Kostrov, X. Immobilization of enzymes on porous silicas–benefits and challenges. Chem. Soc. Rev. 2013, 42, 6277–6289. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Y.; Yang, Y.; Yu, C. Silica-based nanoparticles for enzyme immobilization and delivery. Chem. Asian J. 2022, 17, e202200573. [Google Scholar] [CrossRef]
- Ertesvåg, H. Alginate-modifying enzymes: Biological roles and biotechnological uses. Front. Microbiol. 2015, 6, 523. [Google Scholar] [CrossRef] [PubMed]
- Völzke, J.L.; Shamami, P.H.; Gawlitza, K.; Feldmann, I.; Zimathies, A.; Meyer, K.; Weller, M.G. High-purity corundum as support for affinity extractions from complex samples. Separations 2022, 9, 252. [Google Scholar] [CrossRef]
- Völzke, J.L.; Smatty, S.; Döring, S.; Ewald, S.; Oelze, M.; Fratzke, F.; Flemig, S.; Konthur, Z.; Weller, M.G. Efficient Purification of Polyhistidine-Tagged Recombinant Proteins Using Functionalized Corundum Particles. BioTech 2023, 12, 31. [Google Scholar] [CrossRef]
- Vashist, S.K.; Lam, E.; Hrapovic, S.; Male, K.B.; Luong, J.H. Immobilization of antibodies and enzymes on 3-aminopropyltriethoxysilane-functionalized bioanalytical platforms for biosensors and diagnostics. Chem. Rev. 2014, 114, 11083–11130. [Google Scholar] [CrossRef]
- Sypabekova, M.; Hagemann, A.; Rho, D.; Kim, S. 3-Aminopropyltriethoxysilane (APTES) deposition methods on oxide surfaces in solution and vapor phases for biosensing applications. Biosensors 2022, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 2004, 37, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Clinton, F. Sodium cyanoborohydride—A highly selective reducing agent for organic functional groups. Synthesis 1975, 3, 135–146. [Google Scholar] [CrossRef]
- Germain, P.; Slagmolen, T.; Crichton, R. Relation between stabilization and rigidification of the three-dimensional structure of an enzyme. Biotechnol. Bioeng. 1989, 33, 563–569. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Tsiatsiani, L.; Heck, A.J. Proteomics beyond trypsin. FEBS J. 2015, 282, 2612–2626. [Google Scholar] [CrossRef]
- Sinha, A.; Mann, M. A beginner’s guide to mass spectrometry–based proteomics. Biochemist 2020, 42, 64–69. [Google Scholar] [CrossRef]
- Noor, Z.; Ahn, S.B.; Baker, M.S.; Ranganathan, S.; Mohamedali, A. Mass spectrometry–based protein identification in proteomics—A review. Brief. Bioinform. 2021, 22, 1620–1638. [Google Scholar] [CrossRef]
- Döring, S.; Weller, M.G.; Reinders, Y.; Konthur, Z.; Jaeger, C. Challenges and insights in absolute quantification of recombinant therapeutic antibodies by mass spectrometry: An introductory review. Antibodies 2025, 14, 3. [Google Scholar] [CrossRef]
- Furlong, M.T.; Ouyang, Z.; Wu, S.; Tamura, J.; Olah, T.; Tymiak, A.; Jemal, M. A universal surrogate peptide to enable LC-MS/MS bioanalysis of a diversity of human monoclonal antibody and human Fc-fusion protein drug candidates in pre-clinical animal studies. Biomed. Chromatogr. 2012, 26, 1024–1032. [Google Scholar] [CrossRef]
- Zhang, S.; Jian, W. Recent advances in absolute quantification of peptides and proteins using LC-MS. Rev. Anal. Chem. 2014, 33, 31–47. [Google Scholar] [CrossRef]
- Pappin, D.J.; Hojrup, P.; Bleasby, A.J. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 1993, 3, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Pappin, D.J. Peptide mass fingerprinting using MALDI-TOF mass spectrometry. In Protein Sequencing Protocols; Springer: Berlin/Heidelberg, Germany, 2003; pp. 211–219. [Google Scholar]
- Wu, F.; Zhao, M.; Zhang, Y.; Su, N.; Xiong, Z.; Xu, P. Recombinant acetylated trypsin demonstrates superior stability and higher activity than commercial products in quantitative proteomics studies. Rapid Commun. Mass Spectrom. 2016, 30, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Muriithi, B.; Ippoliti, S.; Finny, A.; Addepalli, B.; Lauber, M. Clean and complete protein digestion with an autolysis resistant trypsin for peptide mapping. J. Proteome Res. 2024, 23, 5221–5228. [Google Scholar] [CrossRef]
- Menneteau, T.; Saveliev, S.; Butré, C.I.; Rivera, A.K.G.; Urh, M.; Delobel, A. Addressing common challenges of biotherapeutic protein peptide mapping using recombinant trypsin. J. Pharm. Biomed. Anal. 2024, 243, 116124. [Google Scholar] [CrossRef]
- Sun, X.; Cai, X.; Wang, R.-Q.; Xiao, J. Immobilized trypsin on hydrophobic cellulose decorated nanoparticles shows good stability and reusability for protein digestion. Anal. Biochem. 2015, 477, 21–27. [Google Scholar] [CrossRef]
- Sahin, S.; Ozmen, I. Covalent immobilization of trypsin on polyvinyl alcohol-coated magnetic nanoparticles activated with glutaraldehyde. J. Pharm. Biomed. Anal. 2020, 184, 113195. [Google Scholar] [CrossRef]
- Aversa, I.F.; Cavalcanti, M.H.; Pereira, T.M.; A. de Castro, A.; Tavano, O.L.; Coelho, Y.L.; da Silva, L.H.; Gorup, L.F.; Ramalho, T.C.; Virtuoso, L.S. Immobilization of Porcine Trypsin in Superparamagnetic Nanoparticles: Enzyme Activity and Stability. ACS Omega 2025, 10, 22970–22983. [Google Scholar] [CrossRef]
- Aslani, E.; Abri, A.; Pazhang, M. Immobilization of trypsin onto Fe3O4@ SiO2–NH2 and study of its activity and stability. Colloids Surf. B Biointerfaces 2018, 170, 553–562. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, S. Immobilization of trypsin from porcine pancreas onto chitosan nonwoven by covalent bonding. Polymers 2019, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
- Miguez, J.P.; Fernandez-Lafuente, R.; Tavano, O.L.; Mendes, A.A. The immobilization and stabilization of trypsin from the porcine pancreas on chitosan and its catalytic performance in protein hydrolysis. Catalysts 2023, 13, 1344. [Google Scholar] [CrossRef]
- Massolini, G.; Calleri, E. Immobilized trypsin systems coupled on-line to separation methods: Recent developments and analytical applications. J. Sep. Sci. 2005, 28, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Regnier, F.E.; Kim, J. Accelerating trypsin digestion: The immobilized enzyme reactor. Bioanalysis 2014, 6, 2685–2698. [Google Scholar] [CrossRef] [PubMed]
- Naldi, M.; Černigoj, U.; Štrancar, A.; Bartolini, M. Towards automation in protein digestion: Development of a monolithic trypsin immobilized reactor for highly efficient on-line digestion and analysis. Talanta 2017, 167, 143–157. [Google Scholar] [CrossRef]
- Reinders, L.M.; Klassen, M.D.; Teutenberg, T.; Jaeger, M.; Schmidt, T.C. Development of a multidimensional online method for the characterization and quantification of monoclonal antibodies using immobilized flow-through enzyme reactors. Anal. Bioanal. Chem. 2021, 413, 7119–7128. [Google Scholar] [CrossRef]
- Fan, X.; Chu, Z.; Zhu, M.; Song, Y.; Zhao, Y.; Meng, B.; Gong, X.; Zhang, D.; Jiang, Y.; Wu, L. Precise control of trypsin immobilization by a programmable DNA tetrahedron designed for ultrafast proteome digestion and accurate protein quantification. Anal. Chem. 2023, 95, 15875–15883. [Google Scholar] [CrossRef]
- Tscheuschner, G.; Schwaar, T.; Weller, M.G. Fast confirmation of antibody identity by MALDI-TOF MS fingerprints. Antibodies 2020, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Tscheuschner, G.; Kaiser, M.N.; Lisec, J.; Beslic, D.; Muth, T.; Krüger, M.; Mages, H.W.; Dorner, B.G.; Knospe, J.; Schenk, J.A. MALDI-TOF-MS-based identification of monoclonal murine anti-SARS-CoV-2 antibodies within one hour. Antibodies 2022, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Schiel, J.E.; Turner, A. The NISTmAb Reference Material 8671 lifecycle management and quality plan. Anal. Bioanal. Chem. 2018, 410, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, E.; Colescott, R.L.; Bossinger, C.D.; Cook, P. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef]
- Sarin, V.K.; Kent, S.B.; Tam, J.P.; Merrifield, R.B. Quantitative monitoring of solid-phase peptide synthesis by the ninhydrin reaction. Anal. Biochem. 1981, 117, 147–157. [Google Scholar] [CrossRef]
- Stauß, A.C.; Fuchs, C.; Jansen, P.; Repert, S.; Alcock, K.; Ludewig, S.; Rozhon, W. The ninhydrin reaction revisited: Optimisation and application for quantification of free amino acids. Molecules 2024, 29, 3262. [Google Scholar] [CrossRef]
- Reinmuth-Selzle, K.; Tchipilov, T.; Backes, A.T.; Tscheuschner, G.; Tang, K.; Ziegler, K.; Lucas, K.; Pöschl, U.; Fröhlich-Nowoisky, J.; Weller, M.G. Determination of the protein content of complex samples by aromatic amino acid analysis, liquid chromatography-UV absorbance, and colorimetry. Anal. Bioanal. Chem. 2022, 414, 4457–4470. [Google Scholar] [CrossRef]
- Erlanger, B.F.; Kokowsky, N.; Cohen, W. The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 1961, 95, 271–278. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Liang, Y.; Yan, X.; Markey, S.P.; Mirokhin, Y.A.; Tchekhovskoi, D.V.; Bukhari, T.H.; Stein, S.E. The NISTmAb tryptic peptide spectral library for monoclonal antibody characterization. MAbs 2018, 10, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J. ABID. Available online: https://github.com/BAMresearch/ABID (accessed on 6 October 2025).
- Schiff, H. Eine neue reihe organischer diamine. Justus Liebigs Ann. Der Chem. 1866, 140, 92–137. [Google Scholar] [CrossRef]
- Wieland, H.; Scheuing, G. Die Fuchsin-schweflige Säure und ihre Farbreaktion mit Aldehyden. Berichte Der Dtsch. Chem. Ges. (A B Ser.) 1921, 54, 2527–2555. [Google Scholar] [CrossRef]
- Hesse, A.; Weller, M.G. Protein quantification by derivatization-free high-performance liquid chromatography of aromatic amino acids. J. Amino Acids 2016, 2016, 7374316. [Google Scholar] [CrossRef]
- Bronsema, K.J.; Bischoff, R.; Pijnappel, W.P.; van der Ploeg, A.T.; Van De Merbel, N.C. Absolute quantification of the total and antidrug antibody-bound concentrations of recombinant human α-glucosidase in human plasma using protein G extraction and LC-MS/MS. Anal. Chem. 2015, 87, 4394–4401. [Google Scholar] [CrossRef]
- Mi, W.; Josephs, R.; Melanson, J.; Dai, X.; Wang, Y.; Zhai, R.; Chu, Z.; Fang, X.; Thibeault, M.; Stocks, B. PAWG pilot study on quantification of SARS-CoV-2 monoclonal antibody-part 1. Metrologia 2022, 59, 08001. [Google Scholar] [CrossRef]
- Martos, G.; Bedu, M.; Josephs, R.; Westwood, S.; Wielgosz, R. Quantification of SARS-CoV-2 monoclonal IgG mass fraction by isotope dilution mass spectrometry. Anal. Bioanal. Chem. 2024, 416, 2423–2437. [Google Scholar] [CrossRef]
- Harries, M.; Smith, I. The development and clinical use of trastuzumab (Herceptin). Endocr. Relat. Cancer 2002, 9, 75–85. [Google Scholar] [CrossRef]
- Lee, B.; Lopez-Ferrer, D.; Kim, B.C.; Na, H.B.; Park, Y.I.; Weitz, K.K.; Warner, M.G.; Hyeon, T.; Lee, S.W.; Smith, R.D. Rapid and efficient protein digestion using trypsin-coated magnetic nanoparticles under pressure cycles. Proteomics 2011, 11, 309–318. [Google Scholar] [CrossRef]
- Bubb, W.A.; Berthon, H.A.; Kuchel, P.W. Tris buffer reactivity with low-molecular-weight aldehydes: NMR characterization of the reactions of glyceraldehyde-3-phosphate. Bioorganic Chem. 1995, 23, 119–130. [Google Scholar] [CrossRef]
- Kim, K.S.; Lee, Y.; Lee, J.H.; Lee, S.S.; Chung, J.M.; Jung, H.S. Optimizing protein crosslinking control: Synergistic quenching effects of glycine, histidine, and lysine on glutaraldehyde reactions. Biochem. Biophys. Res. Commun. 2024, 702, 149567. [Google Scholar] [CrossRef]
- Maciel, J.; Andrad, P.; Neri, D.; Carvalho, L., Jr.; Cardoso, C.; Calazans, G.; Aguiar, J.A.; Silva, M. Preparation and characterization of magnetic levan particles as matrix for trypsin immobilization. J. Magn. Magn. Mater. 2012, 324, 1312–1316. [Google Scholar] [CrossRef]
- Doğan, D.; Sezer, S.; Ulu, A.; Köytepe, S.; Ateş, B. Preparation and characterization of amino-functionalized zeolite/SiO2 materials for trypsin–chymotrypsin co-immobilization. Catal. Lett. 2021, 151, 2463–2477. [Google Scholar] [CrossRef]
- Sanchez, A.; Cruz, J.; Rueda, N.; dos Santos, J.C.; Torres, R.; Ortiz, C.; Villalonga, R.; Fernandez-Lafuente, R. Inactivation of immobilized trypsin under dissimilar conditions produces trypsin molecules with different structures. RSC Adv. 2016, 6, 27329–27334. [Google Scholar] [CrossRef]
- Sun, L.; Zhu, G.; Yan, X.; Mou, S.; Dovichi, N.J. Uncovering immobilized trypsin digestion features from large-scale proteome data generated by high-resolution mass spectrometry. J. Chromatogr. A 2014, 1337, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhang, L.; Zhang, Y. Preparation of high efficiency and low carry-over immobilized enzymatic reactor with methacrylic acid–silica hybrid monolith as matrix for on-line protein digestion. J. Chromatogr. A 2014, 1371, 48–57. [Google Scholar] [CrossRef]
- Kumar, S.; Anderson, K.W. Rapid removal of IgG1 carryover on protease column using protease-safe wash solutions delivered with LC pump for HDX-MS systems. J. Am. Soc. Mass Spectrom. 2024, 36, 340–345. [Google Scholar] [CrossRef]
- Liang, Y.; Tao, D.; Ma, J.; Sun, L.; Liang, Z.; Zhang, L.; Zhang, Y. Hydrophilic monolith based immobilized enzyme reactors in capillary and on microchip for high-throughput proteomic analysis. J. Chromatogr. A 2011, 1218, 2898–2905. [Google Scholar] [CrossRef]
- Kim, B.C.; Lopez-Ferrer, D.; Lee, S.M.; Ahn, H.K.; Nair, S.; Kim, S.H.; Kim, B.S.; Petritis, K.; Camp, D.G.; Grate, J.W. Highly stable trypsin-aggregate coatings on polymer nanofibers for repeated protein digestion. Proteomics 2009, 9, 1893–1900. [Google Scholar] [CrossRef]
- Camperi, J.; Grunert, I.; Heinrich, K.; Winter, M.; Özipek, S.; Hoelterhoff, S.; Weindl, T.; Mayr, K.; Bulau, P.; Meier, M. Inter-laboratory study to evaluate the performance of automated online characterization of antibody charge variants by multi-dimensional LC-MS/MS. Talanta 2021, 234, 122628. [Google Scholar] [CrossRef]
- Kaeek, M.; Khoury, L.R. Bovine Serum Albumin–Trypsin Sponges for Enhanced Enzymatic Stability and Protein Digestion Efficiency. ACS Appl. Bio Mater. 2025, 8, 8889–8902. [Google Scholar] [CrossRef]
- Oezipek, S.; Hoelterhoff, S.; Breuer, S.; Bell, C.; Bathke, A. mD-UPLC-MS/MS: Next generation of mAb characterization by multidimensional ultraperformance liquid chromatography-mass spectrometry and parallel on-column lysC and trypsin digestion. Anal. Chem. 2022, 94, 8136–8145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.-F.; Wang, P.; Han, X.-J.; Qin, T.-T.; Lu, X.; Bai, H.-J. Efficient and rapid digestion of proteins with a dual-enzyme microreactor featuring 3-D pores formed by dopamine/polyethyleneimine/acrylamide-coated KIT-6 molecular sieve. Sci. Rep. 2024, 14, 15667. [Google Scholar] [CrossRef] [PubMed]
- Wilke, M.; Röder, B.; Paul, M.; Weller, M.G. Sintered glass monoliths as supports for affinity columns. Separations 2021, 8, 56. [Google Scholar] [CrossRef]
- Rainer, T.; Egger, A.-S.; Zeindl, R.; Tollinger, M.; Kwiatkowski, M.; Müller, T. 3D-printed high-pressure-resistant immobilized enzyme microreactor (ΜIMER) for protein analysis. Anal. Chem. 2022, 94, 8580–8587. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Döring, S.; Wulfes, B.S.; Atanasova, A.; Jaeger, C.; Walzel, L.; Tscheuschner, G.; Flemig, S.; Gawlitza, K.; Feldmann, I.; Konthur, Z.; et al. Corundum Particles as Trypsin Carrier for Efficient Protein Digestion. BioTech 2026, 15, 2. https://doi.org/10.3390/biotech15010002
Döring S, Wulfes BS, Atanasova A, Jaeger C, Walzel L, Tscheuschner G, Flemig S, Gawlitza K, Feldmann I, Konthur Z, et al. Corundum Particles as Trypsin Carrier for Efficient Protein Digestion. BioTech. 2026; 15(1):2. https://doi.org/10.3390/biotech15010002
Chicago/Turabian StyleDöring, Sarah, Birte S. Wulfes, Aleksandra Atanasova, Carsten Jaeger, Leopold Walzel, Georg Tscheuschner, Sabine Flemig, Kornelia Gawlitza, Ines Feldmann, Zoltán Konthur, and et al. 2026. "Corundum Particles as Trypsin Carrier for Efficient Protein Digestion" BioTech 15, no. 1: 2. https://doi.org/10.3390/biotech15010002
APA StyleDöring, S., Wulfes, B. S., Atanasova, A., Jaeger, C., Walzel, L., Tscheuschner, G., Flemig, S., Gawlitza, K., Feldmann, I., Konthur, Z., & Weller, M. G. (2026). Corundum Particles as Trypsin Carrier for Efficient Protein Digestion. BioTech, 15(1), 2. https://doi.org/10.3390/biotech15010002







