Structural Analysis and Substrate Specificity of D-Carbamoylase from Pseudomonas
Abstract
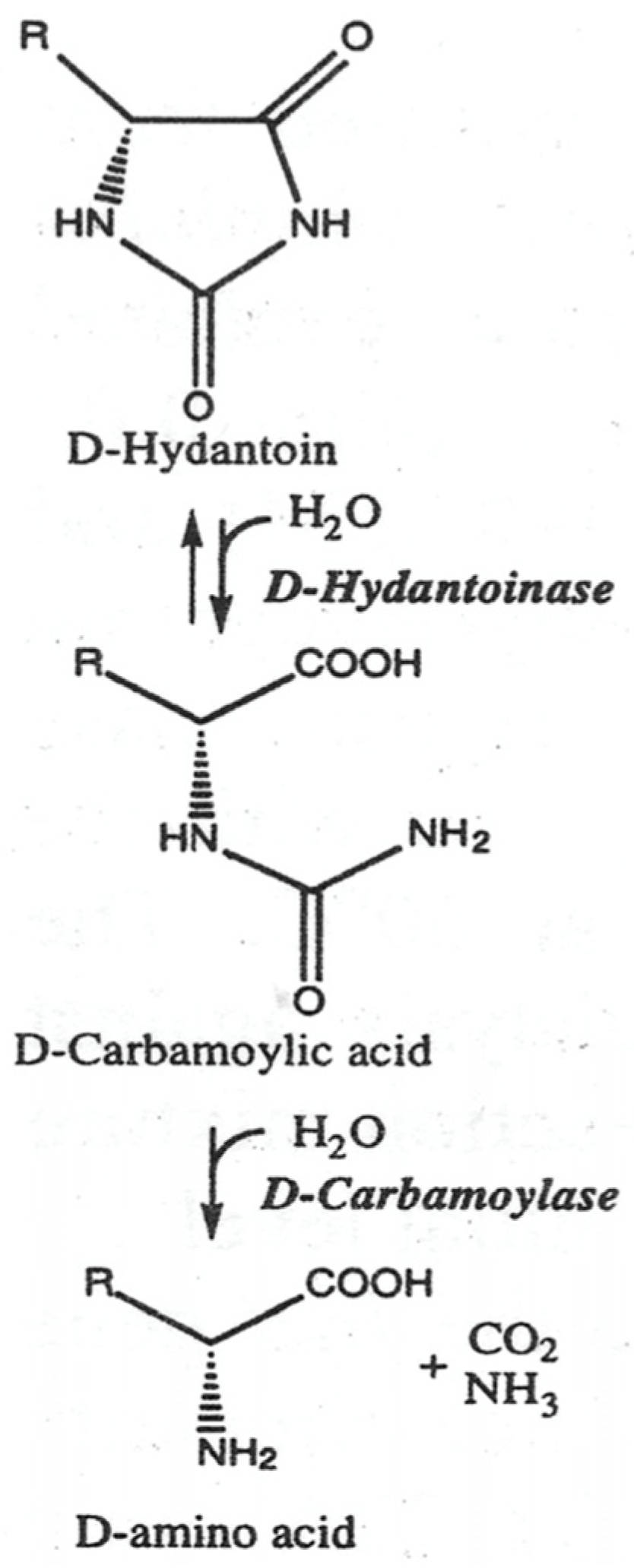
1. Introduction
2. Materials and Methods
2.1. Reagents and Substrates
2.2. Cloning of D-Carbamoylase Gene
2.3. Purification of Recombinant D-Carbamoylase
2.4. D-Carbamoylase Activity Assay
2.5. Solving of Inclusion Bodies
2.6. Protein Assays
2.7. Molecular Docking
2.8. Molecular Dynamics
2.9. English Language Correction
3. Results
3.1. Gene Cloning, D-Carbamoylase Expression and Purification
3.2. The Search of Possible Carbamoylases
3.3. Molecular Docking of D-Carbamoylase
3.4. Molecular Dynamics of D-Carbamoylase Binding to Carbamoylated Amino Acids
3.5. Calculation of Molecular Mechanics of Poisson–Boltzmann Surface Area (Mmpbsa)
3.6. D-Carbamoylase Substrate Specificity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Syldatk, C.; May, O.; Altenbuchner, J.; Mattes, R.; Siemann, M. Microbial hydantoinases—Industrial enzymes from the origin of life? Appl. Microb. Biotechnol. 1999, 51, 293–309. [Google Scholar] [CrossRef] [PubMed]
- Engel, U.; Syldatk, C.; Rudat, J. Stereoselective hydrolysis of aryl-substituted dihydropyrimidines by hydantoinases. Appl. Microb. Biotechnol. 2012, 94, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.J.; Kim, H.S. Optimization of the enzymatic synthesis of D-p-hydroxyphenylglycine from DL-5-substituted hydantoin using D-hydantoinase and N-carbamoylase. Enzyme Microb. Technol. 1995, 17, 63–67. [Google Scholar] [CrossRef]
- Martínez-Rodríguez, S.; Martínez-Gómez, A.I.; Rodríguez-Vico, F.; Clemente-Jiménez, J.M.; Las Heras-Vázquez, F.J. Carbamoylases: Characteristics and applications in biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 85, 441–458. [Google Scholar] [CrossRef]
- Ware, E. The chemistry of the hydantoins. Chem Rev. 1950, 46, 403–470. [Google Scholar] [CrossRef]
- Gaebler, O.H.; Keltch, A.K. On the metabolism of hydantoins and hydantoic acid. J. Biol. Chem. 1926, 70, 763–777. [Google Scholar] [CrossRef]
- Eadie, G.S.; Bernheim, F.; Bernheim, M.L.C. The partial purification and properties of animal and plant hydantoinases. J. Biol. Chem. 1949, 181, 449–458. [Google Scholar] [CrossRef]
- Wallach, D.P.; Grisolia, S. The purification and properties of hydropyrimidine hydrase. J. Biol. Chem. 1957, 226, 277–288. [Google Scholar] [CrossRef]
- Olivieri, R.; Fascetti, E.; Angelini, L.; Degen, L. Enzymatic conversion of N-carbamoyl-D-amino acids to D-amino acids. Enzym. Microb. Tech. 1979, 1, 201–204. [Google Scholar] [CrossRef]
- Möller, A.; Syldatk, C.; Schulze, M. Stereo- and substrate-specificity of a D-hydantoinase and D-N-carbamoyl-amino acid amidohydrolase of Arthrobacter crystallopoietes AM2. Enzyme Microb. Technol. 1988, 10, 618–625. [Google Scholar] [CrossRef]
- Ikenaka, Y.; Nanba, H.; Yamada, Y.; Yajima, K.; Takano, M.; Takahashi, S. Screening, characterization, and cloning of the gene for N-carbamyl-D-amino acid amidohydrolase from thermotolerant soil bacteria. Biosci. Biotechnol. Biochem. 1998, 62, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Chung, M.C.-M.; Hida, S.; Yamada, H.; Shimizu, S. Thermostable N-carbamoyl-D-amino acid amidohydrolase: Screening, purification and characterization. J. Biotechnol. 1994, 38, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Shimizu, S.; Yamada, H. N-carbamoyl-D-amino acid amidohydrolase from Conamonas sp. E222c purification and characterization. Eur. J. Biochem. 1993, 212, 685–691. [Google Scholar] [PubMed]
- Grifantini, R.; Pratesi, C.; Galli, G.; Grandi, G. Topological mapping of the cysteine residues of N-carbamyl-D-amino-acid amidohydrolase and their role in enzymatic activity. J. Biol. Chem. 1996, 271, 9326–9331. [Google Scholar] [CrossRef] [PubMed]
- Nanba, H.; Ikenaka, Y.; Yamda, Y.; Yajima, K.; Takano, M.; Takahashi, S. Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-D-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci. Biotechnol. Biochem. 1998, 62, 875–881. [Google Scholar] [CrossRef]
- Syldatk, C.; Mueller, R.; Siemann, M.; Wagner, F. Microbial and enzymatic production of D-amino acids from DL-5-monosubstituted hydantoins. In Biocatalytic Production of Amino Acids and Derivatives; Rozzell, J.D., Wagner, F., Eds.; Hanser: New York, NY, USA, 1992; pp. 75–127. [Google Scholar]
- Louwrier, A.; Knowles, C.J. The purification and characterization of a novel D(-)-specific carbamoylase enzyme from an Agrobacterium sp. Enzyme Microb. Technol. 1996, 19, 562–571. [Google Scholar] [CrossRef]
- Nozaki, H.; Kira, I.; Watanabe, K.; Yokozeki, K. Purification and properties of D-hydantoin hydrolase and N-car-bamoyl-D-amino acid amidohydrolase from Flavobacterium sp. AJ11199 and Pasteurella sp. AJ11221. J. Mol. Catal B 2005, 32, 205–211. [Google Scholar] [CrossRef]
- Ikenaka, Y.; Nanba, H.; Yajima, K.; Yamada, Y.; Takano, M.; Takahashi, S. Increase in thermostability of N-carbamyl-D-amino acid amidohydrolase on amino acid substitutions. Biosci. Biotechnol. Biochem. 1998, 62, 1668–1671. [Google Scholar] [CrossRef]
- Ikenaka, Y.; Nanba, H.; Yajima, K.; Yamada, Y.; Takano, M.; Takahashi, S. Relationship between an increase in thermostability and amino acid substitutions in N-carbamyl-D-amino acid amidohydrolase. Biosci. Biotechnol. Biochem. 1998, 62, 1672–1675. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Xu, G.; Zhou, J.; Ni, J.; Zhang, L.; Hou, X.; Yin, D.; Rao, Y.; Zhao, Y.-L.; Ni, Y. Structure-guided engineering of D-carbamoylase reveals a key loop at substrate entrance tunnel. ACS Catal. 2020, 10, 12393–12402. [Google Scholar] [CrossRef]
- Nakai, T.; Hasegawa, T.; Yamashita, E.; Yamamoto, M.; Kumasaka, T.; Ueki, T.; Nanba, H.; Ikenaka, Y.; Takahashi, S.; Sato, M.; et al. Crystal structure of N-carbamyl-d-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 2000, 8, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-C.; Hsu, W.-H.; Chien, F.-T.; Chen, C.-Y. Crystal structure and site-directed mutagenesis studies of N-carbamoyl-d-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft. J. Mol. Biol. 2001, 306, 251–261. [Google Scholar] [CrossRef]
- Batisse, N.; Weigel, P.; Lecoq, M.; Sakanyan, V. Two amino acid amidohydrolase genes, encoding L-stereospecific carbamoylase and aminoacylase, are organized into a common operon in Bacillus stearothermophilus. Appl. Environ. Microbiol. 1997, 63, 763–766. [Google Scholar] [CrossRef]
- Weigel, P.; Mark, F.; Aganyants, H.A.; Sakanyan, V.A. Characteristics of thermostable hydantoinases cloned from Geobacilus stearothermophilus. Reports of NAS RA 2013, 113, 92–98. [Google Scholar]
- Aganyants, H.; Weigel, P.; Hovhannisyan, Y.; Lecocq, M.; Koloyan, H.; Hambardzumyan, A.; Hovsepyan, A.; Hallet, J.N.; Sakanyan, V. Rational engineering of the substrate specificity of a thermostable D-hydantoinase (dihydropyrimidinase). High-Throughput 2020, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; p. 2344. [Google Scholar]
- Azevedo, F.; Pereira, H.; Johansson, B. Colony PCR. Methods Mol. Biol. 2017, 1620, 129–139. [Google Scholar] [PubMed]
- Hanczkó, R.; Molnár-Perl, I. Derivatization, stability and chromatographic behavior of ophthaldialdehyde amino acide and amine derivatives: O-phthaldialdehyde/2-mercaptoethanol reagent. Chromatographia 2003, 57, S103–S113. [Google Scholar] [CrossRef]
- Peternel, Š.; Grdadolnik, J.; Gaberc-Porekar, V.; Komel, R. Engineering inclusion bodies for non-denaturing extraction of functional proteins. Microb. Cell. Fact. 2008, 7, 34. [Google Scholar] [CrossRef]
- Peterson, G.L. Determination of total protein. Meth. Enzymol. 1983, 91, 95–119. [Google Scholar]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for multiple sequence alignments. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A Web Compatible Database search or multiple alignment viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1.0: Molecular docking with deep Learning. J. Cheminform. 2021, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Bugnon, M.; Goullieux, M.; Röhrig, U.F.; Perez, M.A.S.; Daina, A.; Michielin, O.; Zoete, V. SwissParam 2023: A modern Web-based tool for efficient small molecule parameterization. J. Chem. Inf. Model. 2023, 63, 6469–6475. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory. Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Corchero, J.L.; Gasser, B.; Resina, D.; Smith, W.; Parrilli, E.; Vazquez, F.; Abasolo, I.; Giuliani, M.; Jäntti, J.; Ferrer, P.; et al. Unconventional microbial systems for the cost-efficient production of high-quality protein therapeutics. Biotechnol Adv. 2013, 31, 140–153. [Google Scholar] [CrossRef]
- Koszagova, R.; Nahalka, J. Inclusion bodies in biotechnology. J. Microbiol. Biotechnol. Food Sci. 2020, 9, 1191–1196. [Google Scholar] [CrossRef]
- Rinas, U.; Garcia-Fruitos, E.; Corchero, J.L.; Vazquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial inclusion bodies: Discovering their better half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Lin, K.Y.; Lu, C.H. Refolding and activation of recombinant N-carbamoyl-D-amino acid amidohydrolase from Escherichia coli inclusion bodies. Process Biochem. 2005, 40, 2135–2141. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Hu, X.; Yang, J. Using native hydantoinase promoter to induce D-carbamoylase soluble expression in Escherichia coli. Biochem. Eng. J. 2008, 41, 12–16. [Google Scholar] [CrossRef]
- Martinez-Alonso, M.; Garcia-Fruitos, E.; Ferrer-Miralles, N.; Rinas, U.; Villaverde, A. Side effects of chaperone gene co-expression in recombinant protein production. Microb. Cell Fact. 2010, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, P.; Saneja, A.; Srichandan, S.; Panda, A.K. Bacterial inclusion bodies: A treasure trove of bioactive proteins. Trends Biotechnol. 2020, 38, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Schein, C.H.; Noteborn, M.H. Formation of soluble recombinant proteins in Escheuichia coli is favored by lower growth temperature. Nat. Biotechnol. 1988, 6, 291–294. [Google Scholar] [CrossRef]
- Chao, Y.-P.; Chiang, C.-J.; Lo, T.E.; Fu, H. Overproduction of D-hydantoinase and carbamoylase in a soluble form in Escherichia coli. Appl. Microbiol. Biotechnol. 2000, 54, 348–353. [Google Scholar] [CrossRef]





| Compound | Specific Activity (U/mg) |
|---|---|
| N-carbamoyl-D-tryptophan | 0.449 ± 0.03 |
| N-carbamoyl-D-phenylalanine | 0.265 ± 0.02 |
| N-carbamoyl-D-alanine | 0.461 ± 0.04 |
| N-carbamoyl-DL-alanine | 0.368 ± 0.03 |
| N-carbamoyl-D-valine | 0.442 ± 0.04 |
| N-carbamoyl-D-leucine | 0.190 ± 0.02 |
| N-carbamoyl-L-tryptophan | 0 |
| N-carbamoyl-L-leucine | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paronyan, M.; Koloyan, H.; Aganyants, H.; Hambardzumyan, A.; Soghomonyan, T.; Avetisyan, S.; Kocharov, S.; Panosyan, H.; Sakanyan, V.; Hovsepyan, A. Structural Analysis and Substrate Specificity of D-Carbamoylase from Pseudomonas. BioTech 2024, 13, 40. https://doi.org/10.3390/biotech13040040
Paronyan M, Koloyan H, Aganyants H, Hambardzumyan A, Soghomonyan T, Avetisyan S, Kocharov S, Panosyan H, Sakanyan V, Hovsepyan A. Structural Analysis and Substrate Specificity of D-Carbamoylase from Pseudomonas. BioTech. 2024; 13(4):40. https://doi.org/10.3390/biotech13040040
Chicago/Turabian StyleParonyan, Marina, Haykanush Koloyan, Hovsep Aganyants, Artur Hambardzumyan, Tigran Soghomonyan, Sona Avetisyan, Sergey Kocharov, Henry Panosyan, Vehary Sakanyan, and Anichka Hovsepyan. 2024. "Structural Analysis and Substrate Specificity of D-Carbamoylase from Pseudomonas" BioTech 13, no. 4: 40. https://doi.org/10.3390/biotech13040040
APA StyleParonyan, M., Koloyan, H., Aganyants, H., Hambardzumyan, A., Soghomonyan, T., Avetisyan, S., Kocharov, S., Panosyan, H., Sakanyan, V., & Hovsepyan, A. (2024). Structural Analysis and Substrate Specificity of D-Carbamoylase from Pseudomonas. BioTech, 13(4), 40. https://doi.org/10.3390/biotech13040040










