Specialized Metabolism of Gordonia Genus: An Integrated Survey on Chemodiversity Combined with a Comparative Genomics-Based Analysis
Abstract
1. Introduction
2. Methods
2.1. PRISMA-Based Literature Collection
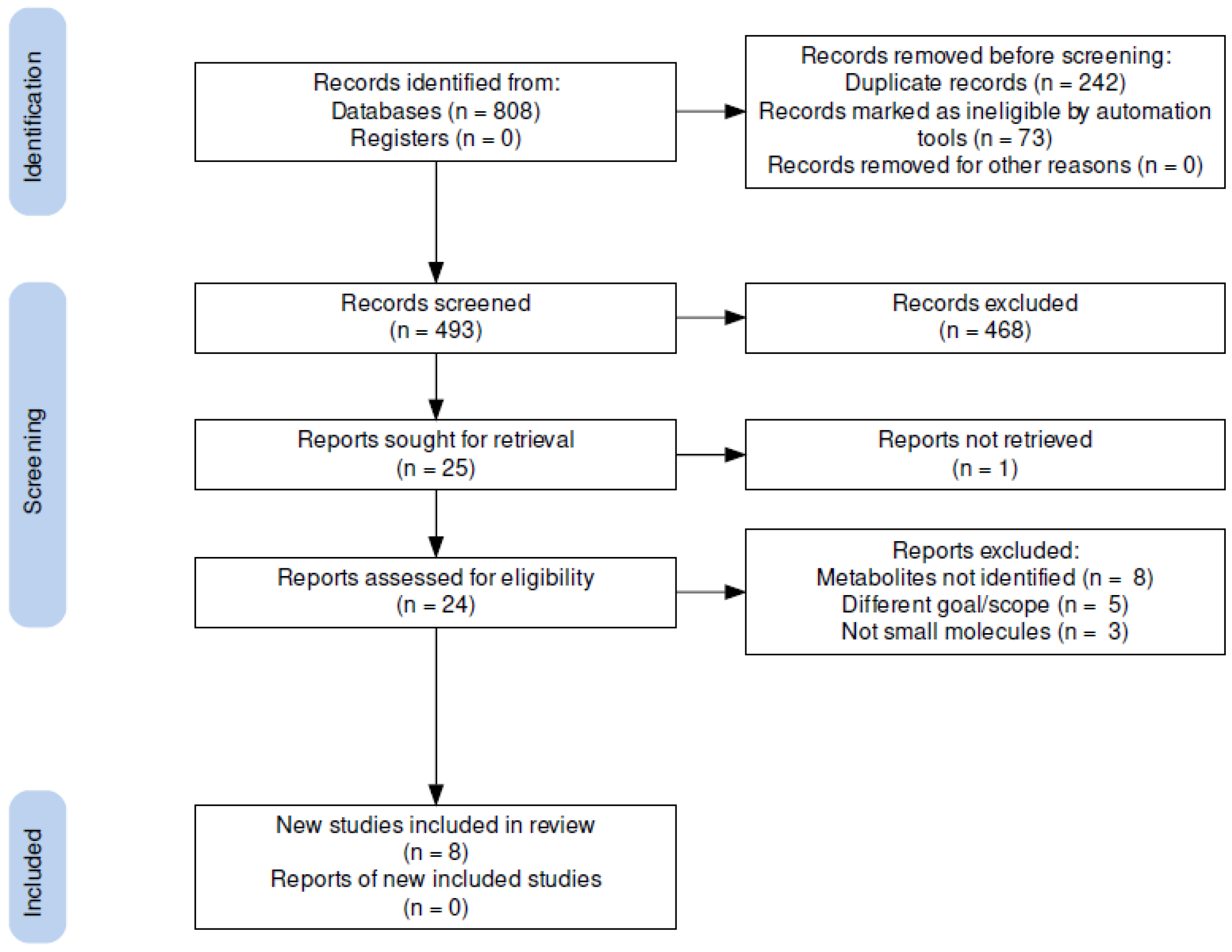
2.1.1. Search Strategy and Study Selection
2.1.2. Data Collection
2.2. Chemoinformatics Analysis
2.3. Comparative Genomics-Based Analysis
2.3.1. Collection of Genome Sequences and Pan-Genome Estimation
2.3.2. Phylogenomic Analysis
2.3.3. BGC Identification and Similarity Comparison
2.4. Data Analysis
3. Results
3.1. General Findings
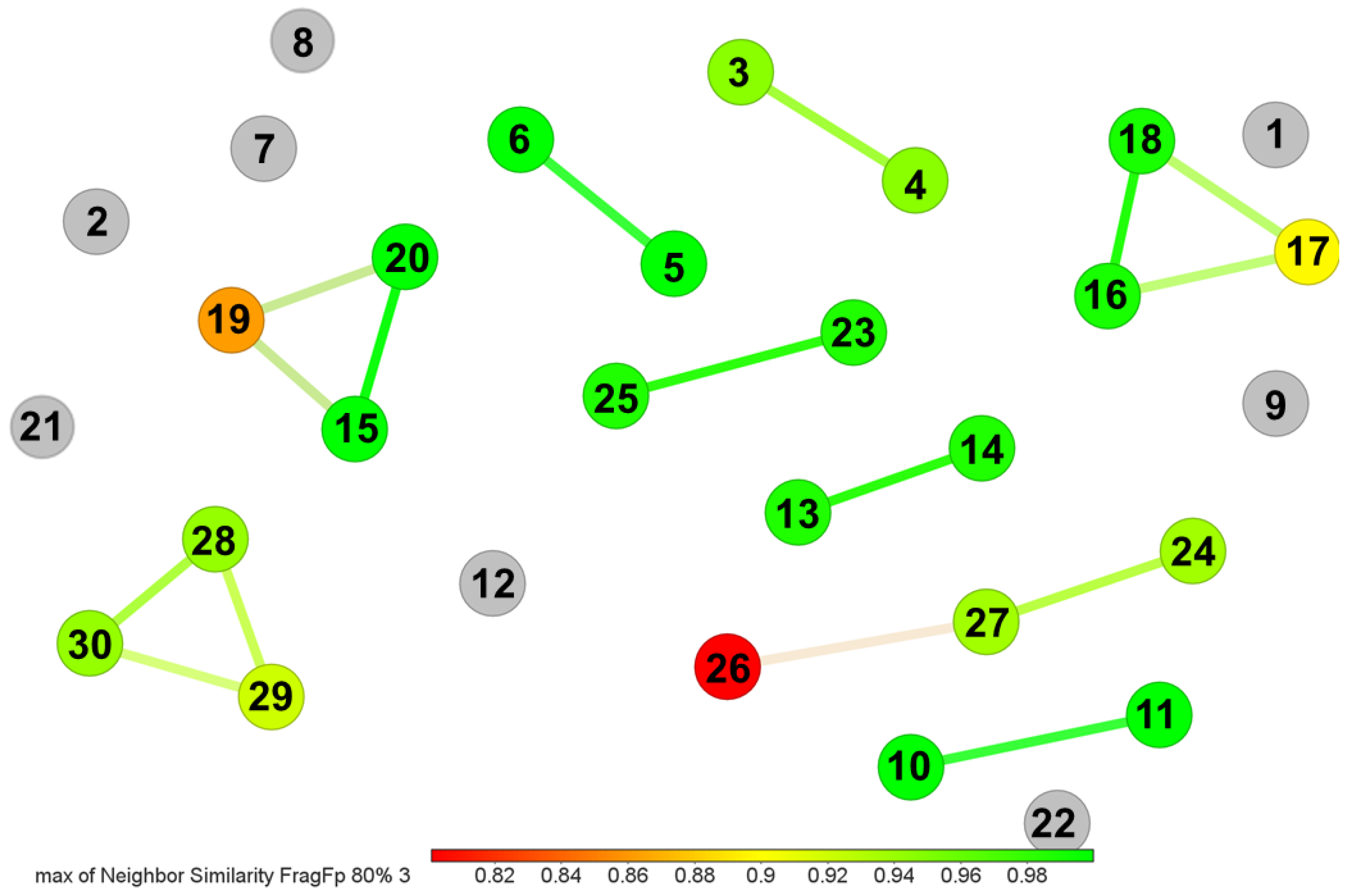
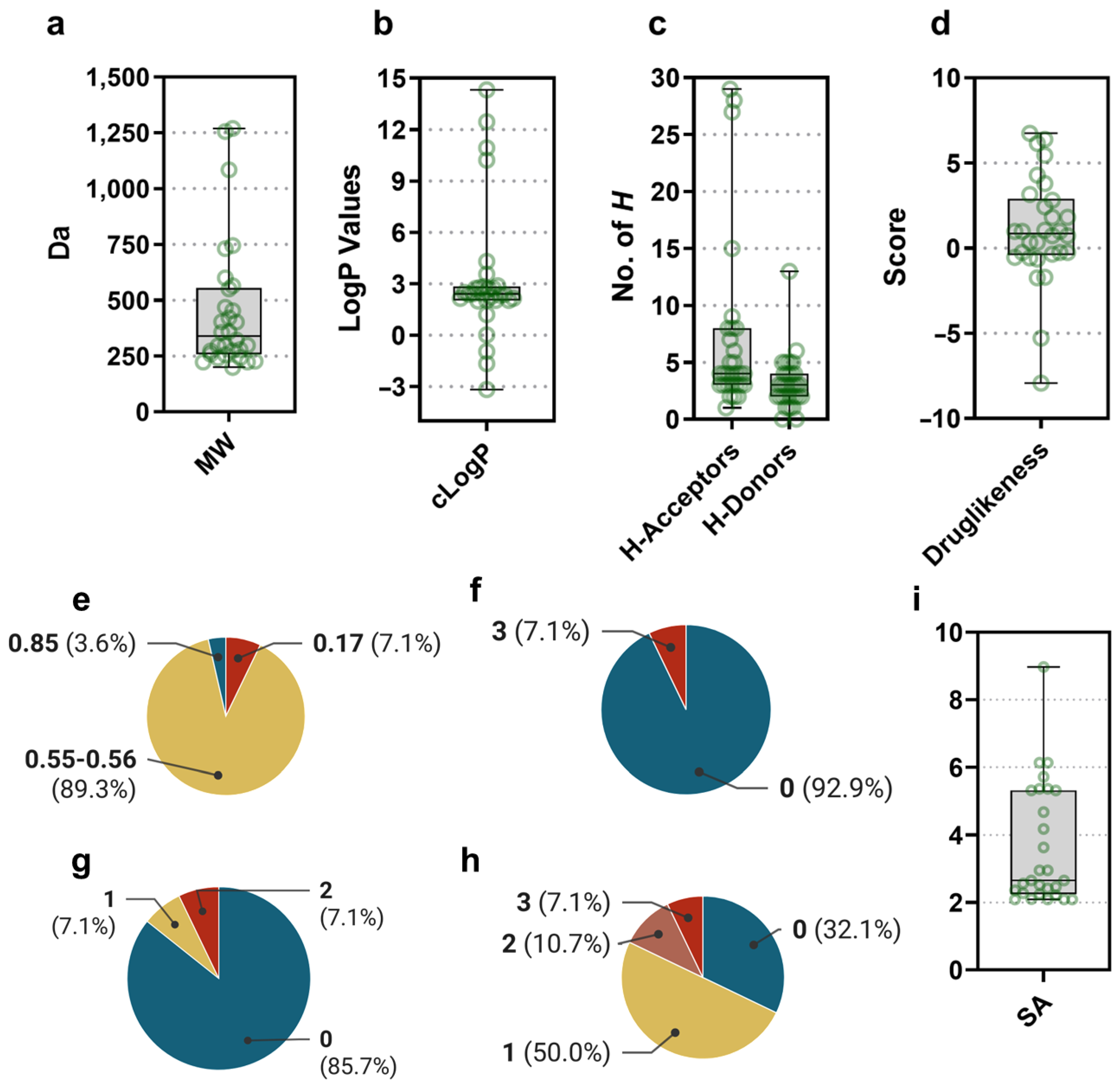
3.2. Gordonia-Derived Metabolites
3.3. Gordonia Pan-Genome
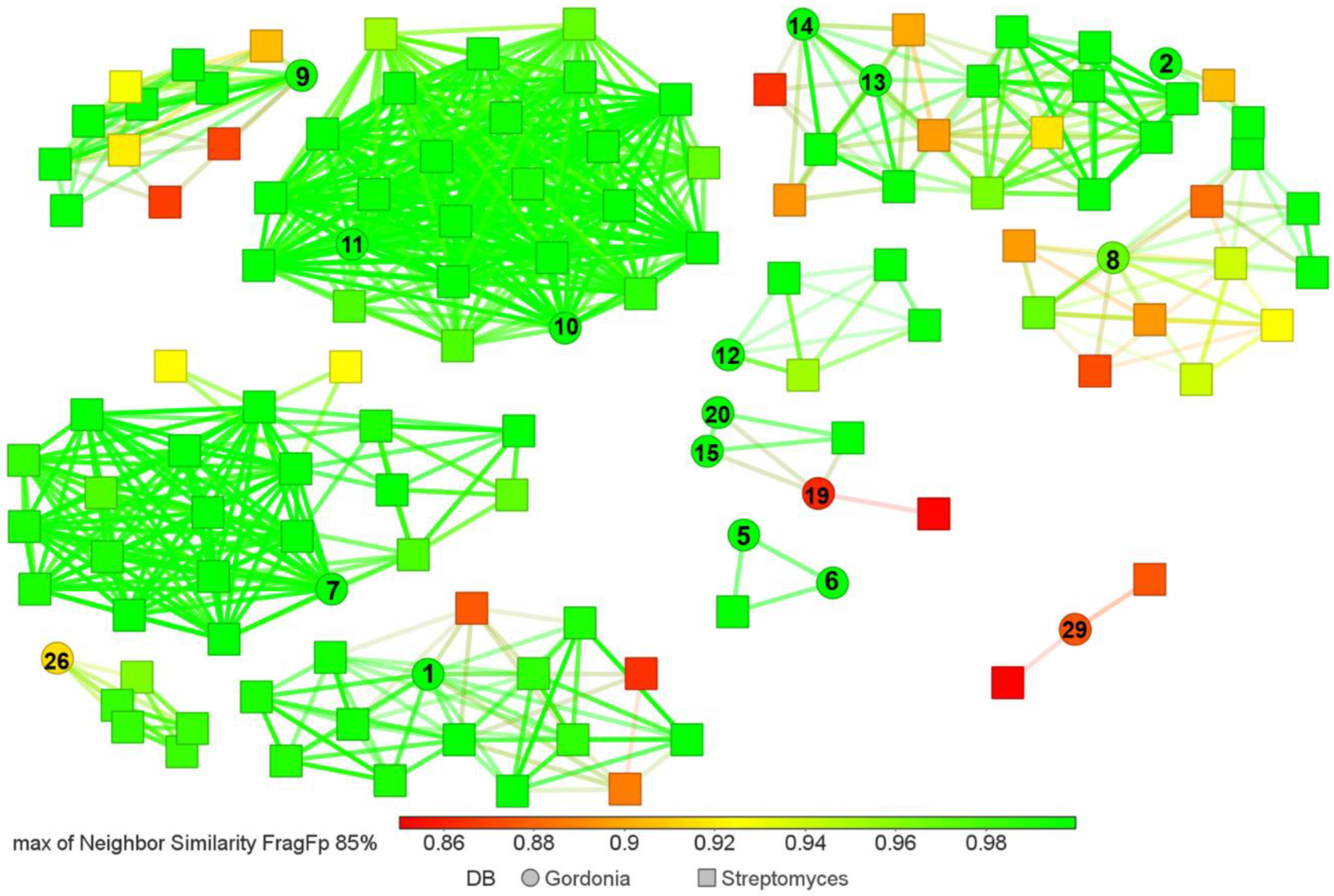
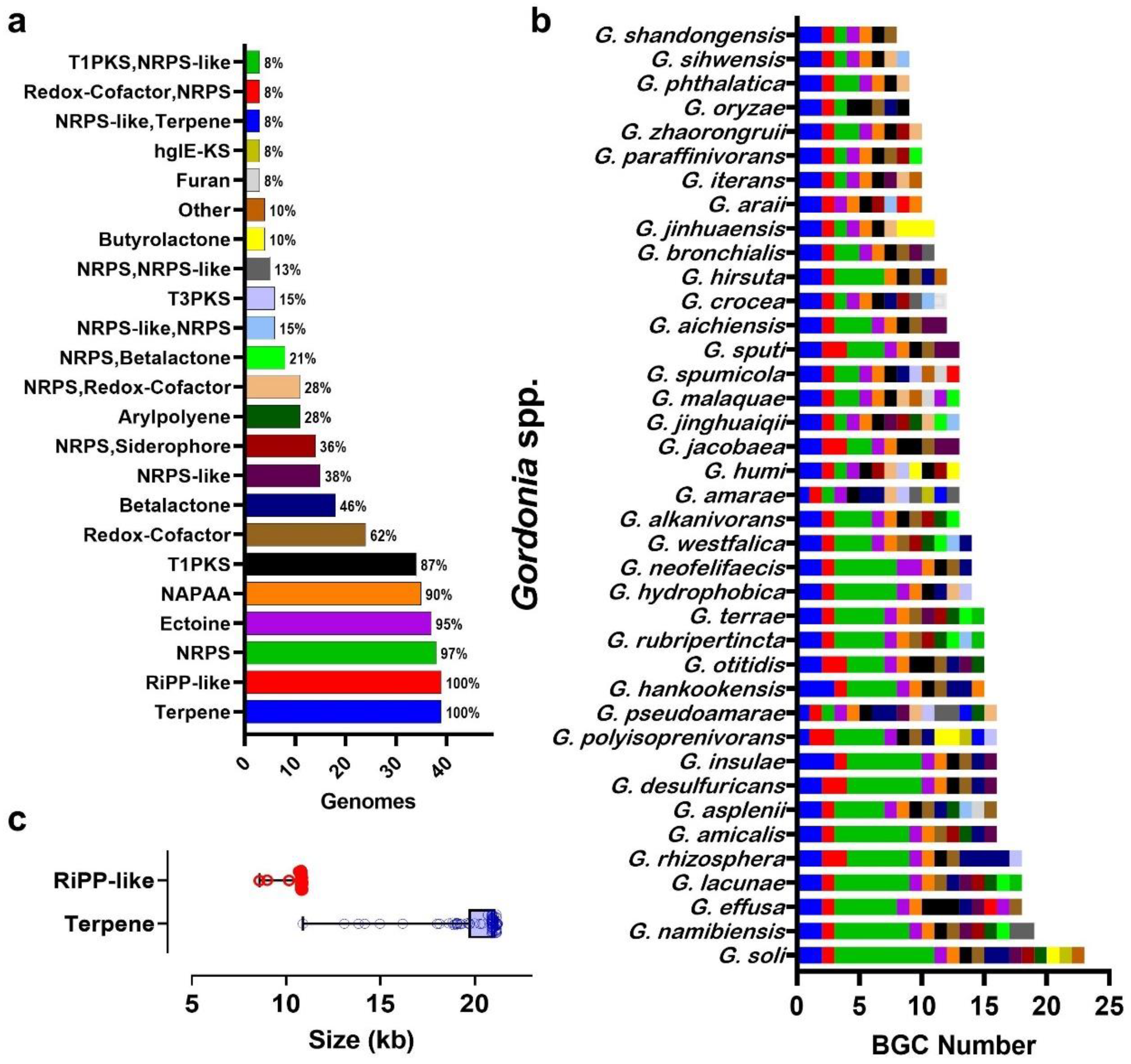
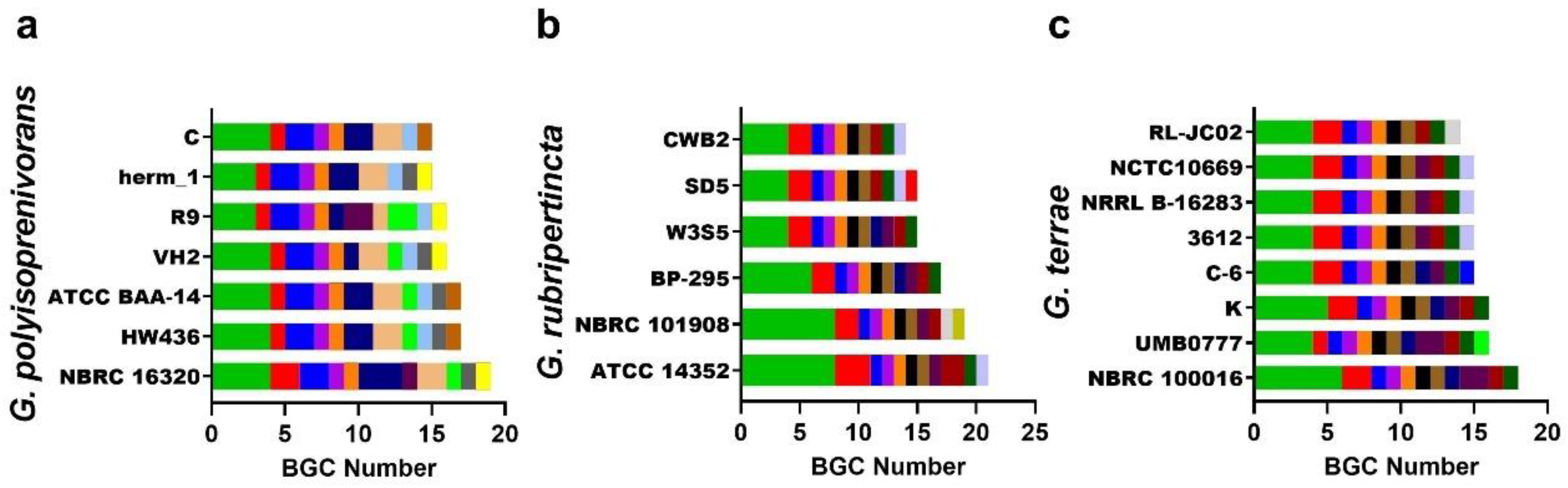
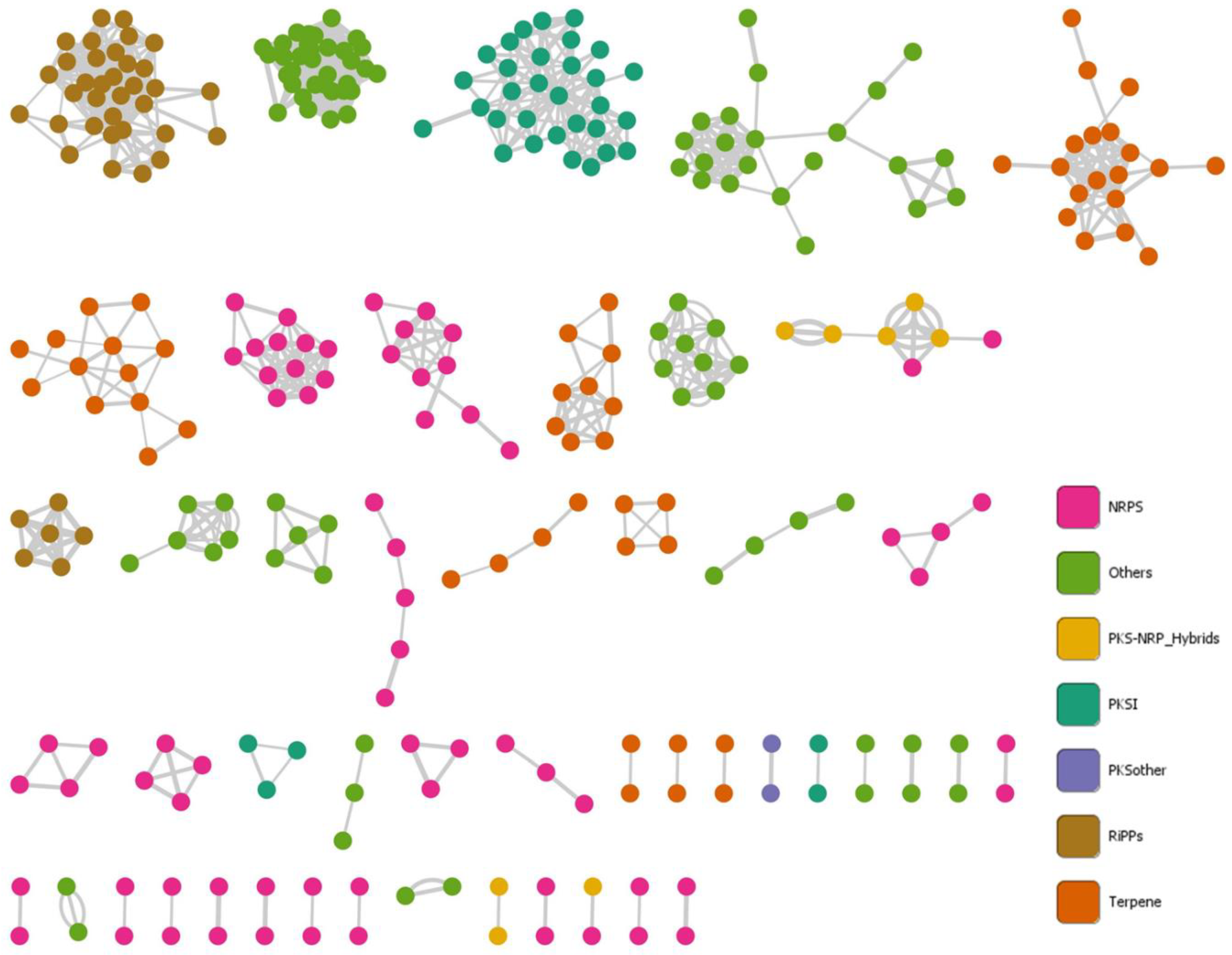
3.4. Gordonia BGC Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wei, B.; Du, A.Q.; Zhou, Z.Y.; Lai, C.; Yu, W.C.; Yu, J.B.; Yu, Y.L.; Chen, J.W.; Zhang, H.W.; Xu, X.W.; et al. An Atlas of Bacterial Secondary Metabolite Biosynthesis Gene Clusters. Environ. Microbiol. 2021, 23, 6981–6992. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A. Streptomyces-Based Cell Factories for Production of Biomolecules and Bioactive Metabolites. In Microbial Cell Factories Engineering for Production of Biomolecules; Elsevier: Amsterdam, The Netherlands, 2021; pp. 183–234. [Google Scholar]
- Sánchez-Suárez, J.; Coy-Barrera, E.; Villamil, L.; Díaz, L. Streptomyces-Derived Metabolites with Potential Photoprotective Properties—A Systematic Literature Review and Meta-Analysis on the Reported Chemodiversity. Molecules 2020, 25, 3221. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-S.; Kim, H.-J.; Lee, H.-S.; Kim, P.; Kim, E.-S. Genome Mining of Rare Actinomycetes and Cryptic Pathway Awakening. Process Biochem. 2015, 50, 1184–1193. [Google Scholar] [CrossRef]
- Jose, P.A.; Sivakala, K.K.; Jha, B. Non-Streptomyces Actinomycetes and Natural Products: Recent Updates. In Studies in Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 395–409. [Google Scholar]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Smida, J.; Collins, M.D. Evidence of Phylogenetic Heterogeneity within the Genus Rhodococcus: Revival of the Genus Gordona (Tsukamura). J. Gen. Appl. Microbiol. 1988, 34, 341–348. [Google Scholar] [CrossRef]
- Arenskötter, M.; Bröker, D.; Steinbüchel, A. Biology of the Metabolically Diverse Genus Gordonia. Appl. Environ. Microbiol. 2004, 70, 3195–3204. [Google Scholar] [CrossRef]
- Andalibi, F.; Fatahi-Bafghi, M. Gordonia: Isolation and Identification in Clinical Samples and Role in Biotechnology. Folia Microbiol. 2017, 62, 245–252. [Google Scholar] [CrossRef]
- Sowani, H.; Kulkarni, M.; Zinjarde, S. An Insight into the Ecology, Diversity and Adaptations of Gordonia Species. Crit. Rev. Microbiol. 2018, 44, 393–413. [Google Scholar] [CrossRef]
- O’Brien, P.A.; Webster, N.S.; Miller, D.J.; Bourne, D.G. Host-Microbe Coevolution: Applying Evidence from Model Systems to Complex Marine Invertebrate Holobionts. MBio 2019, 10. [Google Scholar] [CrossRef]
- Voser, T.M.; Campbell, M.D.; Carroll, A.R. How Different Are Marine Microbial Natural Products Compared to Their Terrestrial Counterparts? Nat. Prod. Rep. 2022, 39, 7–19. [Google Scholar] [CrossRef]
- Hug, J.J.; Krug, D.; Müller, R. Bacteria as Genetically Programmable Producers of Bioactive Natural Products. Nat. Rev. Chem. 2020, 4, 172–193. [Google Scholar] [CrossRef]
- Rosero-Chasoy, G.; Rodríguez-Jasso, R.M.; Aguilar, C.N.; Buitrón, G.; Chairez, I.; Ruiz, H.A. Microbial Co-Culturing Strategies for the Production High Value Compounds, a Reliable Framework towards Sustainable Biorefinery Implementation—An Overview. Bioresour. Technol. 2021, 321, 124458. [Google Scholar] [CrossRef] [PubMed]
- Verstraete, W.; Yanuka-Golub, K.; Driesen, N.; De Vrieze, J. Engineering Microbial Technologies for Environmental Sustainability: Choices to Make. Microb. Biotechnol. 2022, 15, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- McCauley, E.P.; Piña, I.C.; Thompson, A.D.; Bashir, K.; Weinberg, M.; Kurz, S.L.; Crews, P. Highlights of Marine Natural Products Having Parallel Scaffolds Found from Marine-Derived Bacteria, Sponges, and Tunicates. J. Antibiot. 2020, 73, 504–525. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Twardowski, T.; Wohlgemuth, R. Bioeconomy for Sustainable Development. Biotechnol. J. 2019, 14, 1800638. [Google Scholar] [CrossRef]
- Akinsemolu, A.A. The Role of Microorganisms in Achieving the Sustainable Development Goals. J. Clean. Prod. 2018, 182, 139–155. [Google Scholar] [CrossRef]
- Haines, A. Health in the Bioeconomy. Lancet Planet. Heal. 2021, 5, e4–e5. [Google Scholar] [CrossRef]
- Wohlgemuth, R.; Twardowski, T.; Aguilar, A. Bioeconomy Moving forward Step by Step—A Global Journey. N. Biotechnol. 2021, 61, 22–28. [Google Scholar] [CrossRef]
- Timmis, K.; de Lorenzo, V.; Verstraete, W.; Ramos, J.L.; Danchin, A.; Brüssow, H.; Singh, B.K.; Timmis, J.K. The Contribution of Microbial Biotechnology to Economic Growth and Employment Creation. Microb. Biotechnol. 2017, 10, 1137–1144. [Google Scholar] [CrossRef]
- Neergheen-Bhujun, V.; Awan, A.T.; Baran, Y.; Bunnefeld, N.; Chan, K.; dela Cruz, T.E.; Egamberdieva, D.; Elsässer, S.; Johnson, M.V.; Komai, S.; et al. Biodiversity, Drug Discovery, and the Future of Global Health: Introducing the Biodiversity to Biomedicine Consortium, a Call to Action. J. Glob. Health 2017, 7, 020304. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Chevrette, M.G.; Gavrilidou, A.; Mantri, S.; Selem-Mojica, N.; Ziemert, N.; Barona-Gómez, F. The Confluence of Big Data and Evolutionary Genome Mining for the Discovery of Natural Products. Nat. Prod. Rep. 2021, 38, 2024–2040. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene Cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Soldatou, S.; Eldjarn, G.H.; Huerta-Uribe, A.; Rogers, S.; Duncan, K.R. Linking Biosynthetic and Chemical Space to Accelerate Microbial Secondary Metabolite Discovery. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef]
- Drzyzga, O. The Strengths and Weaknesses of Gordonia: A Review of an Emerging Genus with Increasing Biotechnological Potential. Crit. Rev. Microbiol. 2012, 38, 300–316. [Google Scholar] [CrossRef]
- Rethlefsen, M.L.; Kirtley, S.; Waffenschmidt, S.; Ayala, A.P.; Moher, D.; Page, M.J.; Koffel, J.B. PRISMA-S: An Extension to the PRISMA Statement for Reporting Literature Searches in Systematic Reviews. Syst. Rev. 2021, 10, 39. [Google Scholar] [CrossRef]
- Aghaei Chadegani, A.; Salehi, H.; Md Yunus, M.M.; Farhadi, H.; Fooladi, M.; Farhadi, M.; Ale Ebrahim, N. A Comparison between Two Main Academic Literature Collections: Web of Science and Scopus Databases. Asian Soc. Sci. 2013, 9, 18–26. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Caicedo-Montoya, C.; Manzo-Ruiz, M.; Ríos-Estepa, R. Pan-genome of the genus Streptomyces and prioritization of biosynthetic gene clusters with potential to produce antibiotic compounds. Front. Microbiol. 2021, 12, 677558. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative Genomics: The Bacterial Pan-Genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Costa, S.S.; Guimarães, L.C.; Silva, A.; Soares, S.C.; Baraúna, R.A. First Steps in the Analysis of Prokaryotic Pan-Genomes. Bioinform. Biol. Insights 2020, 14. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Los Alamos National Laboratory Gap Strip/Squeeze v2.1.0. Available online: http://www.hiv.lanl.gov/content/sequence/GAPSTREEZE/gap.html (accessed on 23 January 2022).
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Hodcroft, E.; Hué, S.; Fearnhill, E.; Delpech, V.; Brown, A.J.L.; Lycett, S. Automated Analysis of Phylogenetic Clusters. BMC Bioinform. 2013, 14, 317. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Haddaway, N.R.; Pritchard, C.C.; McGuinness, L.A. PRISMA2020: R Package and ShinyApp for Producing PRISMA 2020 Compliant Flow Diagrams, Version 0.0.2. 2021. Available online: http://irep.ntu.ac.uk/id/eprint/43397/ (accessed on 2 October 2022).
- De Miguel, T.; Sieiro, C.; Poza, M.; Villa, T.G. Analysis of Canthaxanthin and Related Pigments from Gordonia Jacobaea Mutants. J. Agric. Food Chem. 2001, 49, 1200–1202. [Google Scholar] [CrossRef]
- Schwabe, R.; Senges, C.H.R.; Bandow, J.E.; Heine, T.; Lehmann, H.; Wiche, O.; Schlömann, M.; Levicán, G.; Tischler, D. Cultivation Dependent Formation of Siderophores by Gordonia Rubripertincta CWB2. Microbiol. Res. 2020, 238, 126481. [Google Scholar] [CrossRef]
- Schneider, K.; Graf, E.; Irran, E.; Nicholson, G.; Stainsby, F.M.; Goodfellow, M.; Borden, S.A.; Keller, S.; Süssmuth, R.D.; Fiedler, H.P. Bendigoles A∼C, New Steroids from Gordonia Australis Acta 2299. J. Antibiot. 2008, 61, 356–364. [Google Scholar] [CrossRef]
- Park, H.B.; Park, J.-S.; Lee, S.I.; Shin, B.; Oh, D.-C.; Kwon, H.C. Gordonic Acid, a Polyketide Glycoside Derived from Bacterial Coculture of Streptomyces and Gordonia Species. J. Nat. Prod. 2017, 80, 2542–2546. [Google Scholar] [CrossRef]
- Takaichi, S.; Maoka, T.; Akimoto, N.; Carmona, M.L.; Yamaoka, Y. Carotenoids in a Corynebacterineae, Gordonia Terrae AIST-1: Carotenoid Glucosyl Mycoloyl Esters. Biosci. Biotechnol. Biochem. 2008, 72, 2615–2622. [Google Scholar] [CrossRef]
- Lin, Z.; Marett, L.; Hughen, R.W.; Flores, M.; Forteza, I.; Ammon, M.A.; Concepcion, G.P.; Espino, S.; Olivera, B.M.; Rosenberg, G.; et al. Neuroactive Diol and Acyloin Metabolites from Cone Snail-Associated Bacteria. Bioorganic Med. Chem. Lett. 2013, 23, 4867–4869. [Google Scholar] [CrossRef]
- Shamikh, Y.I.; El Shamy, A.A.; Gaber, Y.; Abdelmohsen, U.R.; Madkour, H.A.; Horn, H.; Hassan, H.M.; Elmaidomy, A.H.; Alkhalifah, D.H.M.; Hozzein, W.N. Actinomycetes from the Red Sea Sponge Coscinoderma Mathewsi: Isolation, Diversity, and Potential for Bioactive Compounds Discovery. Microorganisms 2020, 8, 783. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xu, M.; Liu, H.; Yu, T.; Guo, P.; Liu, W.; Jin, X. Antimicrobial Compounds Were Isolated from the Secondary Metabolites of Gordonia, a Resident of Intestinal Tract of Periplaneta Americana. AMB Express 2021, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A. Secondary Metabolic Gene Clusters: Evolutionary Toolkits for Chemical Innovation. Trends Genet. 2010, 26, 449–457. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An Updated Compendium of Streptomycetes Natural Products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.P.; Hersey, A.; Montanari, D.; Overington, J. Probing the Links between in vitro Potency, ADMET and Physicochemical Parameters. Nat. Rev. Drug Discov. 2011, 10, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Gohil, N.; Bhattacharjee, G.; Singh, V. An Introduction to Microbial Cell Factories for Production of Biomolecules. In Microbial Cell Factories Engineering for Production of Biomolecules; Elsevier: Amsterdam, The Netherlands, 2021; pp. 1–19. [Google Scholar]
- Abdel-Razek, A.S.; El-Naggar, M.E.; Allam, A.; Morsy, O.M.; Othman, S.I. Microbial Natural Products in Drug Discovery. Processes 2020, 8, 470. [Google Scholar] [CrossRef]
- O’Brien, J.; Wright, G.D. An Ecological Perspective of Microbial Secondary Metabolism. Curr. Opin. Biotechnol. 2011, 22, 552–558. [Google Scholar] [CrossRef]
- Conti, R.; Chagas, F.O.; Caraballo-Rodriguez, A.M.; da Paixão Melo, W.G.; do Nascimento, A.M.; Cavalcanti, B.C.; de Moraes, M.O.; Pessoa, C.; Costa-Lotufo, L.V.; Krogh, R.; et al. Endophytic Actinobacteria from the Brazilian Medicinal Plant Lychnophora Ericoides Mart. and the Biological Potential of Their Secondary Metabolites. Chem. Biodivers. 2016, 13, 727–736. [Google Scholar] [CrossRef]
- Kim, H.-K.; Jeong, M.-J.; Kong, M.-Y.; Han, M.Y.; Son, K.-H.; Kim, H.M.; Hong, S.H.; Kwon, B.-M. Inhibition of Shc/Grb2 Protein–Protein Interaction Suppresses Growth of B104-1-1 Tumors Xenografted in Nude Mice. Life Sci. 2005, 78, 321–328. [Google Scholar] [CrossRef]
- Rodrigues, L.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential Applications in Medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kang, J.Y.; Hong, Y.K.; Baek, H.H.; Shin, H.W.; Kim, M.S. Isolation and Structural Determination of the Antifouling Diketopiperazines from Marine-Derived Streptomyces Praecox 291-11. Biosci. Biotechnol. Biochem. 2012, 76, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Karthik, L. Pharmacology of FDA-Approved Medicines from Actinobacteria. In Actinobacteria; Springer: Singapore, 2022; pp. 265–276. [Google Scholar]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an Inexhaustible Source of Naturally Occurring Antibiotics. Antibiotics 2018, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Poorinmohammad, N.; Bagheban-Shemirani, R.; Hamedi, J. Genome Mining for Ribosomally Synthesised and Post-Translationally Modified Peptides (RiPPs) Reveals Undiscovered Bioactive Potentials of Actinobacteria. Antonie Van Leeuwenhoek 2019, 112, 1477–1499. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic Gene Clusters and the Evolution of Fungal Chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef] [PubMed]




| Gordonia Strain | Isolation Features | Natural Product Type | Bioactivity Potential | Ref. b | |
|---|---|---|---|---|---|
| Habitat a | Country | ||||
| G. jacobaea CECT 5282 | TFL | Spain | Extract | N/A c | [49] |
| G. rubripertincta CWB2 (DSM 46758) | TFL | Germany | Compound | N/A | [50] |
| G. australis Acta 2299 | FWFL | Australia | Compound | Steroid receptors binding | [51] |
| Gordonia sp. KMC005 | FWFL | Republic of Korea | Compound | Antimicrobial | [52] |
| G. terrae AIST-1 | MFL | Japan | Compound | N/A | [53] |
| Gordonia sp. 647 W.R.1a.05 | MHA | Philippines | Compound | Neuroactive | [54] |
| Gordonia sp. UA19 | MHA | Egypt | Extract | Antimicrobial | [55] |
| G. terrae WA 4-31 | THA | China | Compound | Antimicrobial, Cytotoxic | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Suárez, J.; Díaz, L.; Coy-Barrera, E.; Villamil, L. Specialized Metabolism of Gordonia Genus: An Integrated Survey on Chemodiversity Combined with a Comparative Genomics-Based Analysis. BioTech 2022, 11, 53. https://doi.org/10.3390/biotech11040053
Sánchez-Suárez J, Díaz L, Coy-Barrera E, Villamil L. Specialized Metabolism of Gordonia Genus: An Integrated Survey on Chemodiversity Combined with a Comparative Genomics-Based Analysis. BioTech. 2022; 11(4):53. https://doi.org/10.3390/biotech11040053
Chicago/Turabian StyleSánchez-Suárez, Jeysson, Luis Díaz, Ericsson Coy-Barrera, and Luisa Villamil. 2022. "Specialized Metabolism of Gordonia Genus: An Integrated Survey on Chemodiversity Combined with a Comparative Genomics-Based Analysis" BioTech 11, no. 4: 53. https://doi.org/10.3390/biotech11040053
APA StyleSánchez-Suárez, J., Díaz, L., Coy-Barrera, E., & Villamil, L. (2022). Specialized Metabolism of Gordonia Genus: An Integrated Survey on Chemodiversity Combined with a Comparative Genomics-Based Analysis. BioTech, 11(4), 53. https://doi.org/10.3390/biotech11040053









