Gut Microbiome Remains Static in Functional Abdominal Pain Disorders Patients Compared to Controls: Potential for Diagnostic Tools
Abstract
1. Introduction
2. Materials and Methods
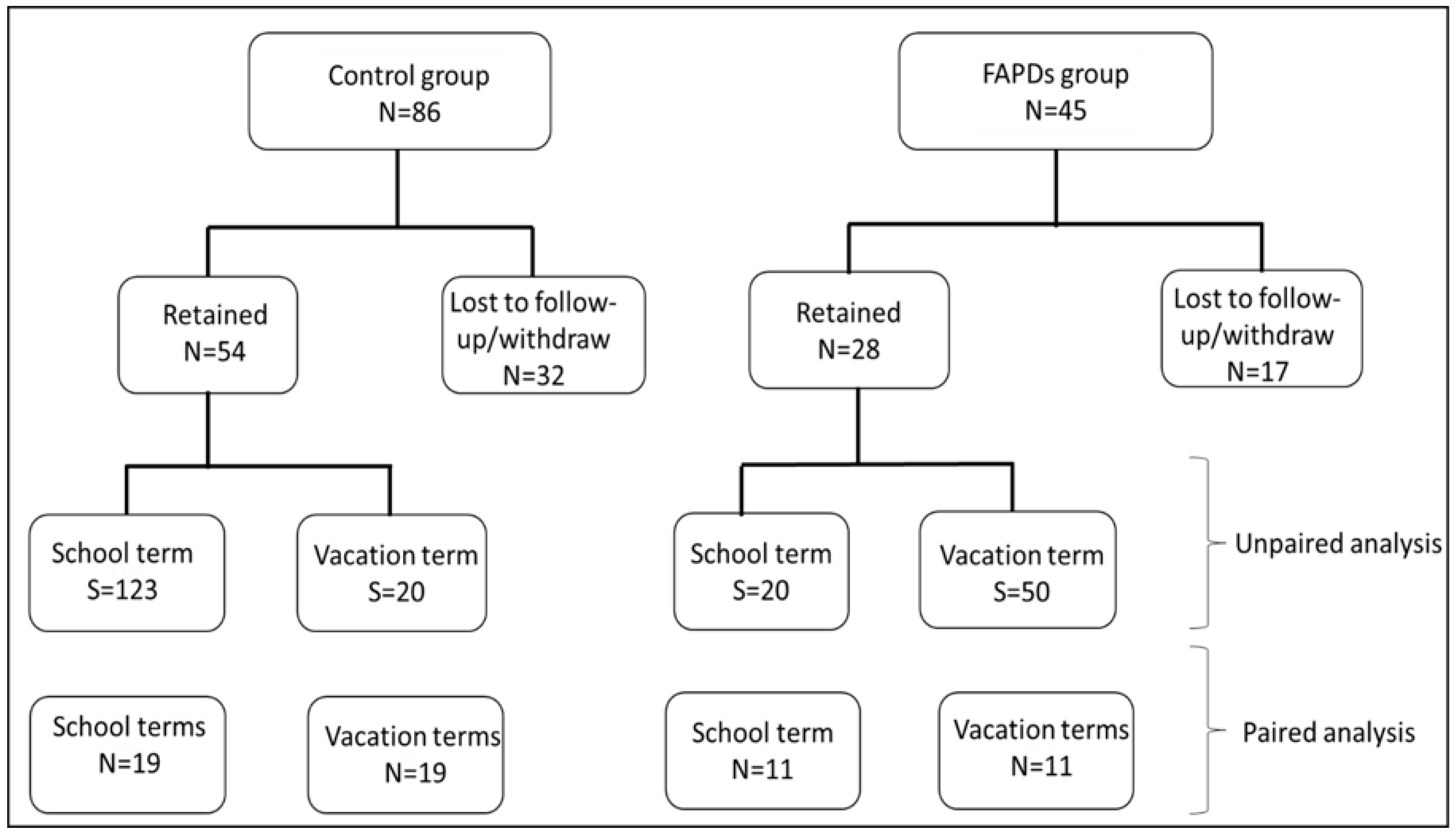
2.1. Recruitment and Study Design
2.2. Stool Samples Collection and Sequencing
2.3. Statistical Analyses
3. Results
3.1. Participants’ Demographics
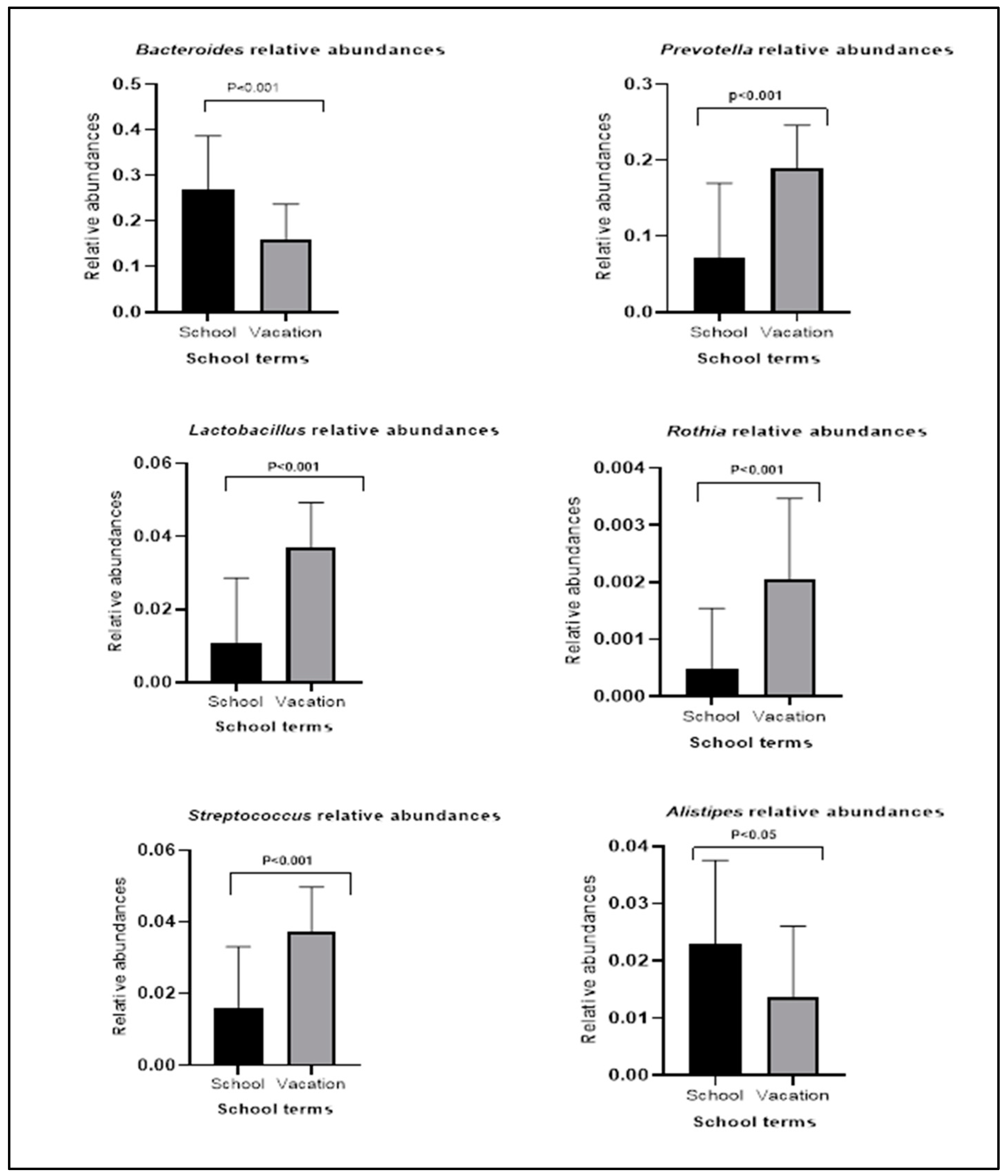
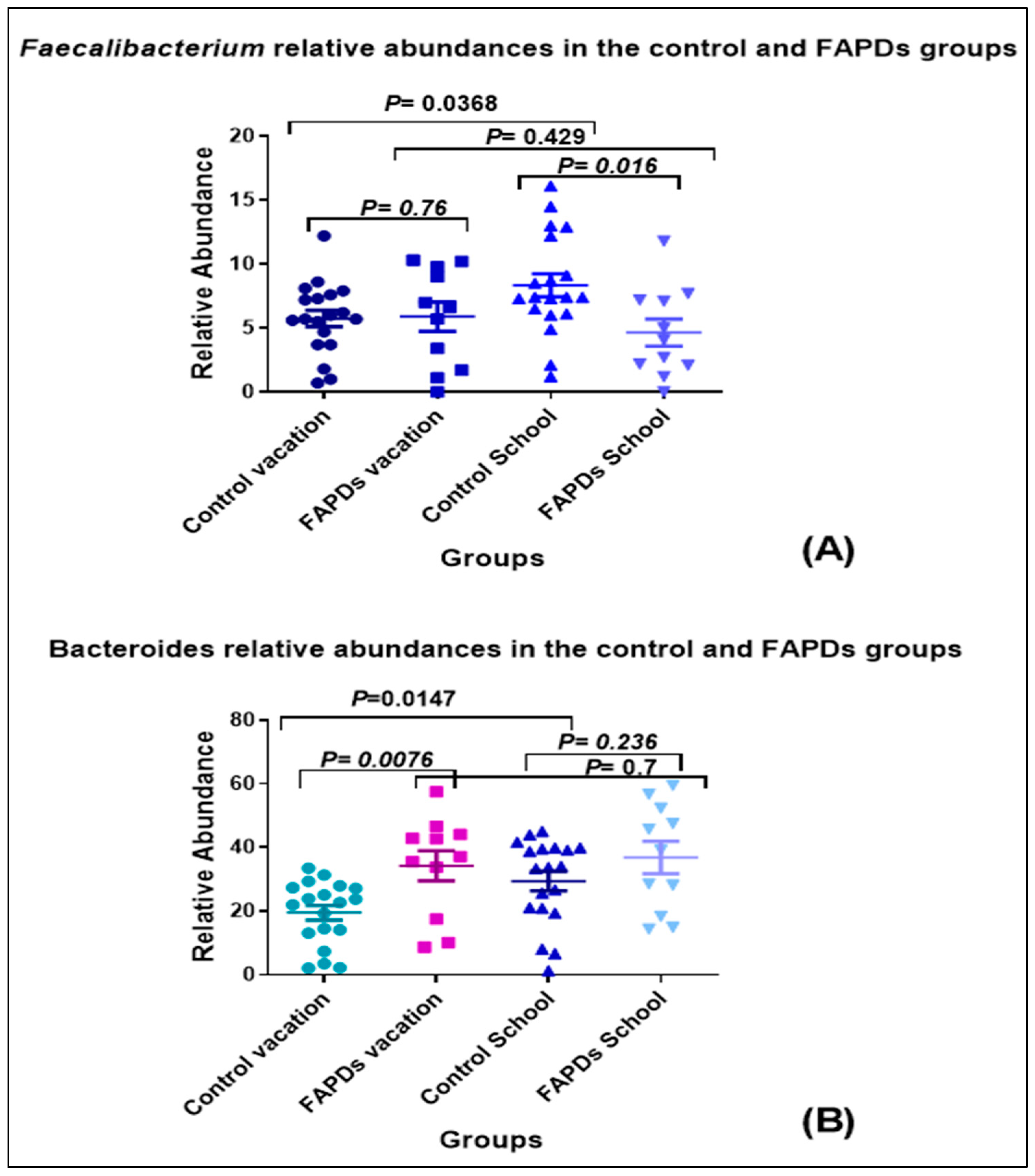
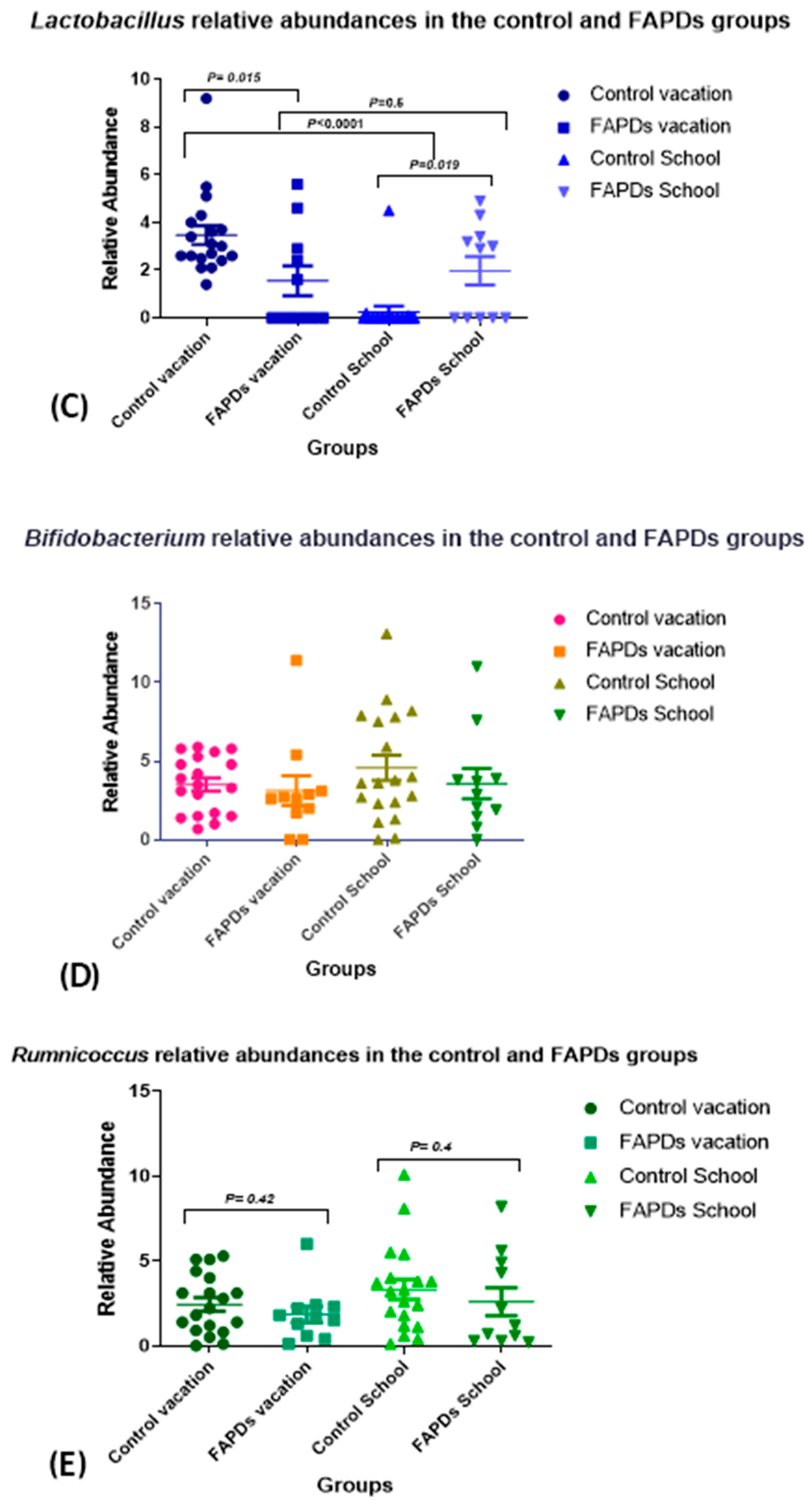
3.2. Bacterial Relative Abundances within Groups during School Terms and Vacation
3.3. Alpha and Beta Diversity Comparison in FAPDs and Control Groups
4. Discussion
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saps, M.; Seshadri, R.; Sztainberg, M.; Schaffer, G.; Marshall, B.M.; Di Lorenzo, C. A prospective school-based study of abdominal pain and other common somatic complaints in children. J. Pediatr. 2009, 154, 322–326. [Google Scholar] [CrossRef]
- Hyams, J.S.; Di Lorenzo, C.; Saps, M.; Shulman, R.J.; Staiano, A.; van Tilburg, M. Functional Disorders: Children and Adolescents. Gastroenterology 2016, 150, 1456–1468.e2. [Google Scholar] [CrossRef] [PubMed]
- Langshaw, A.H.; Rosen, J.M.; Pensabene, L.; Borrelli, O.; Salvatore, S.; Thapar, N.; Concolino, D.; Saps, M. Overlap between functional abdominal pain disorders and organic diseases in children. Rev. Gastroenterol. Mex. 2018, 83, 268–274. [Google Scholar] [CrossRef]
- Shankar, V.; Agans, R.; Holmes, B.; Raymer, M.; Paliy, O. Do gut microbial communities differ in pediatric IBS and health? Gut Microbes 2013, 4, 347–352. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chumpitazi, B.P.; Hollister, E.B.; Oezguen, N.; Tsai, C.M.; McMeans, A.R.; Luna, R.A.; Savidge, T.C.; Versalovic, J.; Shulman, R.J. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes 2014, 5, 165–175. [Google Scholar] [CrossRef]
- Abomoelak, B.; Pemberton, V.; Deb, C.; Campion, S.; Vinson, M.; Mauck, J.; Manipadam, J.; Sudakaran, S.; Patel, S.; Saps, M.; et al. The Gut Microbiome Alterations in Pediatric Patients with Functional Abdominal Pain Disorders. Microorganisms 2021, 9, 2354. [Google Scholar] [CrossRef]
- Saps, M.; Blank, C.; Khan, S.; Seshadri, R.; Marshall, B.; Bass, L.; Di Lorenzo, C. Seasonal variation in the presentation of abdominal pain. J. Pediatr. Gastroenterol. Nutr. 2008, 46, 279–284. [Google Scholar] [CrossRef]
- Saps, M.; Hudgens, S.; Mody, R.; Lasch, K.; Harikrishnan, V.; Baum, C. Seasonal patterns of abdominal pain consultations among adults and children. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 290–296. [Google Scholar] [CrossRef]
- Saps, M.; Adams, P.; Bonilla, S.; Chogle, A.; Nichols-Vinueza, D. Parental report of abdominal pain and abdominal pain-related functional gastrointestinal disorders from a community survey. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 707–710. [Google Scholar] [CrossRef]
- Peter, J.; Fournier, C.; Durdevic, M.; Knoblich, L.; Keip, B.; Dejaco, C.; Trauner, M.; Moser, G. A Microbial Signature of Psychological Distress in Irritable Bowel Syndrome. Psychosom. Med. 2018, 80, 698–709. [Google Scholar] [CrossRef]
- Michels, N.; Van de Wiele, T.; Fouhy, F.; O’Mahony, S.; Clarke, G.; Keane, J. Gut microbiome patterns depending on children’s psychosocial stress: Reports versus biomarkers. Brain Behav. Immun. 2019, 80, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Schmid-Zalaudek, K.; Fredriksen, P.M.; Fabiani, E.; Fredriksen, M.V.; Brix, B.; Rossler, A.; Jantscher, A.; Goswami, N. Children’s hair cortisol and hair testosterone concentrations in response to environmental changes: From summer holidays to school. Stress 2021, 24, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.S.; Gupta, A. Influence of Early Life, Diet, and the Environment on the Microbiome. Clin. Gastroenterol. Hepatol. 2019, 17, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Matenchuk, B.A.; Mandhane, P.J.; Kozyrskyj, A.L. Sleep, circadian rhythm, and gut microbiota. Sleep Med. Rev. 2020, 53, 101340. [Google Scholar] [CrossRef]
- Young, L.; Kemper, K.J. Integrative care for pediatric patients with pain. J. Altern. Complement. Med. 2013, 19, 627–632. [Google Scholar] [CrossRef]
- Edwards, T.; Friesen, C.; Schurman, J.V. Classification of pediatric functional gastrointestinal disorders related to abdominal pain using Rome III vs. Rome IV criterions. BMC Gastroenterol. 2018, 18, 41. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef]
- Dominianni, C.; Wu, J.; Hayes, R.B.; Ahn, J. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol. 2014, 14, 103. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Steenwyk, J.L.; Rokas, A. ggpubfigs: Colorblind-Friendly Color Palettes and ggplot2 Graphic System Extensions for Publication-Quality Scientific Figures. Microbiol. Resour. Announc. 2021, 10, e0087121. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, C.M.; Frank, D.N.; Williamson, K.; Arbet, J.; Wagner, B.D.; Kechris, K.; Kroehl, M.E. tidyMicro: A pipeline for microbiome data analysis and visualization using the tidyverse in R. BMC Bioinform. 2021, 22, 41. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Shiny-phyloseq: Web application for interactive microbiome analysis with provenance tracking. Bioinformatics 2015, 31, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Maharshak, N.; Ringel, Y.; Katibian, D.; Lundqvist, A.; Sartor, R.B.; Carroll, I.M.; Ringel-Kulka, T. Fecal and Mucosa-Associated Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Dig. Dis. Sci. 2018, 63, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Derrien, M.; Tornblom, H.; Brazeilles, R.; Cools-Portier, S.; Dore, J.; Storsrud, S.; Le Neve, B.; Ohman, L.; Simren, M. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology 2017, 152, 111–123.e8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhi, F. Lower Level of Bacteroides in the Gut Microbiota Is Associated with Inflammatory Bowel Disease: A Meta-Analysis. Biomed. Res. Int. 2016, 2016, 5828959. [Google Scholar] [CrossRef]
- Srinivasan, R.; Akobeng, A.K. Thalidomide and thalidomide analogues for induction of remission in Crohn’s disease. Cochrane Database Syst. Rev. 2009, CD007350. [Google Scholar] [CrossRef]
- Chakraborti, C.K. New-found link between microbiota and obesity. World J. Gastrointest. Pathophysiol. 2015, 6, 110–119. [Google Scholar] [CrossRef]
- Jeffery, I.B.; O’Toole, P.W.; Ohman, L.; Claesson, M.J.; Deane, J.; Quigley, E.M.; Simren, M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012, 61, 997–1006. [Google Scholar] [CrossRef]
- Bauer, P.V.; Hamr, S.C.; Duca, F.A. Regulation of energy balance by a gut-brain axis and involvement of the gut microbiota. Cell. Mol. Life Sci. 2015, 73, 737–755. [Google Scholar] [CrossRef]
- Agnello, M.; Carroll, L.N.; Imam, N.; Pino, R.; Palmer, C.; Varas, I.; Greene, C.; Hitschfeld, M.; Gupta, S.; Almonacid, D.E.; et al. Gut microbiome composition and risk factors in a large cross-sectional IBS cohort. BMJ Open Gastroenterol. 2020, 7, e000345. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. The Gut-Brain Axis and the Microbiome: Clues to Pathophysiology and Opportunities for Novel Management Strategies in Irritable Bowel Syndrome (IBS). J. Clin. Med. 2018, 7, 6. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Lu, P.L.; Blom, P.J.J.; Qian, Q.; Velasco-Benitez, C.A.; Benninga, M.A.; Saps, M. Colombian School Children With Functional Gastrointestinal Disorders Respond Differently to Family Stress Than Healthy Children. J. Pediatr. Gastroenterol. Nutr. 2019, 68, e58–e61. [Google Scholar] [CrossRef] [PubMed]
- Deleu, S.; Machiels, K.; Raes, J.; Verbeke, K.; Vermeire, S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Sumitani, K.; Tokunaga, M.; Ishihara, N.; Okubo, T.; Fujisawa, T. Consumption of partially hydrolysed guar gum stimulates Bifidobacteria and butyrate-producing bacteria in the human large intestine. Benef. Microbes 2015, 6, 451–455. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martin, R.; Bermudez-Humaran, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef]
- Tana, C.; Umesaki, Y.; Imaoka, A.; Handa, T.; Kanazawa, M.; Fukudo, S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol. Motil. 2010, 22, 512-e115. [Google Scholar] [CrossRef]
- Xiao, L.; Liu, Q.; Luo, M.; Xiong, L. Gut Microbiota-Derived Metabolites in Irritable Bowel Syndrome. Front. Cell. Infect. Microbiol. 2021, 11, 729346. [Google Scholar] [CrossRef]
- Xu, C.; Jia, Q.; Zhang, L.; Wang, Z.; Zhu, S.; Wang, X.; Liu, Y.; Li, M.; Zhang, J.; Wang, X.; et al. Multiomics Study of Gut Bacteria and Host Metabolism in Irritable Bowel Syndrome and Depression Patients. Front. Cell. Infect. Microbiol. 2020, 10, 580980. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gong, J.; Wang, W.; Long, Y.; Fu, X.; Fu, Y.; Qian, W.; Hou, X. Are there any different effects of Bifidobacterium, Lactobacillus and Streptococcus on intestinal sensation, barrier function and intestinal immunity in PI-IBS mouse model? PLoS ONE 2014, 9, e90153. [Google Scholar] [CrossRef] [PubMed]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Sanchez, M.I.; Hall, G.B.; Ghajar, K.; Nardelli, A.; Bolino, C.; Lau, J.T.; Martin, F.P.; Cominetti, O.; Welsh, C.; Rieder, A.; et al. Probiotic Bifidobacterium longum NCC3001 Reduces Depression Scores and Alters Brain Activity: A Pilot Study in Patients With Irritable Bowel Syndrome. Gastroenterology 2017, 153, 448–459.e8. [Google Scholar] [CrossRef] [PubMed]


| Rome IV Classification | Number of Patients (N = 23) |
|---|---|
| Irritable Bowel Syndrome | 15 |
| Functional Dyspepsia | 13 |
| Abdominal Migraines | 7 |
| Functional Constipation | 5 |
| Adolescent Rumination Syndrome | 2 |
| Functional Abdominal Pain—nos | 1 |
| Functional Nausea | 1 |
| Functional Vomiting | 1 |
| Cyclic Vomiting Syndrome | 1 |
| Non-retentive Fecal Incontinence | 0 |
| Aerophagia | 0 |
| Feature | Coefficient | N | N Not 0 | p Value | Q Value |
|---|---|---|---|---|---|
| Bulleidia | 0.020327637 | 142 | 38 | 3.42E-11 | 8.05E-09 |
| Lactobacillus | 0.132172676 | 142 | 66 | 1.19E-09 | 1.35E-07 |
| Granulicatella | 0.048838228 | 142 | 59 | 2.15E-09 | 1.35E-07 |
| Fusobacterium | 0.050860561 | 142 | 60 | 2.29E-09 | 1.35E-07 |
| Alloprevotella | 0.051864263 | 142 | 52 | 4.50E-09 | 2.12E-07 |
| Neisseria | 0.069465138 | 142 | 58 | 5.68E-09 | 2.23E-07 |
| Atopobium | 0.051192828 | 142 | 53 | 1.89E-08 | 5.39E-07 |
| Solobacterium | 0.030782911 | 142 | 52 | 1.94E-08 | 5.39E-07 |
| Porphyromonas | 0.047079679 | 142 | 62 | 2.23E-08 | 5.39E-07 |
| Klebsiella | 0.071987738 | 142 | 59 | 2.29E-08 | 5.39E-07 |
| Moraxella | 0.033278381 | 142 | 51 | 2.59E-08 | 5.53E-07 |
| Oribacterium | 0.042789854 | 142 | 46 | 3.34E-08 | 6.40E-07 |
| Peptostreptococcus | 0.053773359 | 142 | 53 | 3.54E-08 | 6.40E-07 |
| Rothia | 0.029322402 | 142 | 37 | 3.93E-08 | 6.60E-07 |
| Enterobacteriaceae;__ | 0.066945366 | 142 | 100 | 4.47E-08 | 7.01E-07 |
| Stenotrophomonas | 0.049565018 | 142 | 52 | 5.54E-08 | 8.14E-07 |
| Veillonella | 0.088241544 | 142 | 90 | 6.25E-08 | 8.64E-07 |
| Peptoniphilus | 0.047833579 | 142 | 53 | 1.03E-07 | 1.34E-06 |
| Prevotella | 0.265054965 | 142 | 90 | 2.68E-07 | 3.31E-06 |
| Parvimonas | 0.029579179 | 142 | 56 | 3.53E-07 | 4.14E-06 |
| Alloscardovia | 0.048653236 | 142 | 52 | 5.22E-07 | 5.85E-06 |
| Streptococcus | 0.083962421 | 142 | 133 | 6.72E-07 | 7.18E-06 |
| Megasphaera | 0.04917445 | 142 | 61 | 9.65E-07 | 9.86E-06 |
| Treponema | 0.009954054 | 142 | 23 | 1.35E-06 | 1.32E-05 |
| Gemella | 0.042810564 | 142 | 52 | 1.53E-06 | 1.43E-05 |
| Muribaculaceae | 0.014753749 | 142 | 33 | 2.51E-06 | 2.27E-05 |
| Leptotrichia | 0.015595742 | 142 | 36 | 2.61E-06 | 2.27E-05 |
| Unassigned | 0.019034108 | 142 | 41 | 3.07E-06 | 2.58E-05 |
| Candidatus_Saccharimonas | 0.0167842 | 142 | 27 | 4.47E-06 | 3.62E-05 |
| Aggregatibacter | 0.021560817 | 142 | 28 | 4.62E-06 | 3.62E-05 |
| Saccharimonadaceae;g__TM7x | 0.02284891 | 142 | 64 | 1.06E-05 | 8.01E-05 |
| Actinomyces | 0.033553175 | 142 | 48 | 1.21E-05 | 8.87E-05 |
| Stomatobaculum | 0.021394494 | 142 | 41 | 1.50E-05 | 0.000106562 |
| g__Lachnoanaerobaculum | 0.00686581 | 142 | 7 | 1.55E-05 | 0.000107388 |
| g__Mitochondria | 0.003454846 | 142 | 6 | 4.88E-05 | 0.000327673 |
| Campylobacter | 0.018845676 | 142 | 55 | 5.96E-05 | 0.000388877 |
| Bergeyella | 0.004791259 | 142 | 12 | 6.85E-05 | 0.000434922 |
| Bacilli;__;__;__ | 0.025746385 | 142 | 43 | 7.79E-05 | 0.000481685 |
| Haemophilus | 0.036289019 | 142 | 84 | 9.31E-05 | 0.000561025 |
| Holdemania | −0.017748516 | 142 | 87 | 0.000133085 | 0.000781876 |
| Selenomonas | 0.006838253 | 142 | 13 | 0.00022944 | 0.001315086 |
| Bacteroides | −0.133075069 | 142 | 142 | 0.000238368 | 0.001332788 |
| Capnocytophaga | 0.010475722 | 142 | 26 | 0.000243872 | 0.001332788 |
| Collinsella | −0.036662689 | 142 | 82 | 0.000279701 | 0.00149386 |
| [Eubacterium]_nodatum_group | 0.016523165 | 142 | 47 | 0.000393628 | 0.00205561 |
| Lachnospiraceae;__ | −0.034771487 | 142 | 137 | 0.002178163 | 0.011127572 |
| Absconditabacteriales_(SR1) | 0.005121259 | 142 | 19 | 0.002877646 | 0.014388232 |
| Pseudomonas | 0.001975578 | 142 | 3 | 0.005612792 | 0.027479295 |
| Fusicatenibacter | −0.028887826 | 142 | 134 | 0.006120786 | 0.029354791 |
| Incertae_Sedis | −0.024059973 | 142 | 139 | 0.006365965 | 0.029920034 |
| Enterococcus | −0.016408838 | 142 | 38 | 0.007243531 | 0.033377053 |
| Alistipes | −0.036479155 | 142 | 135 | 0.008722811 | 0.039420396 |
| Ochrobactrum | 0.004726509 | 142 | 21 | 0.009257382 | 0.041046884 |
| _Family_XIII_AD3011_group | −0.017485629 | 142 | 87 | 0.010148283 | 0.044163825 |
| Anaeroglobus | 0.005091565 | 142 | 13 | 0.011408078 | 0.048743606 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abomoelak, B.; Saps, M.; Sudakaran, S.; Deb, C.; Mehta, D. Gut Microbiome Remains Static in Functional Abdominal Pain Disorders Patients Compared to Controls: Potential for Diagnostic Tools. BioTech 2022, 11, 50. https://doi.org/10.3390/biotech11040050
Abomoelak B, Saps M, Sudakaran S, Deb C, Mehta D. Gut Microbiome Remains Static in Functional Abdominal Pain Disorders Patients Compared to Controls: Potential for Diagnostic Tools. BioTech. 2022; 11(4):50. https://doi.org/10.3390/biotech11040050
Chicago/Turabian StyleAbomoelak, Bassam, Miguel Saps, Sailendharan Sudakaran, Chirajyoti Deb, and Devendra Mehta. 2022. "Gut Microbiome Remains Static in Functional Abdominal Pain Disorders Patients Compared to Controls: Potential for Diagnostic Tools" BioTech 11, no. 4: 50. https://doi.org/10.3390/biotech11040050
APA StyleAbomoelak, B., Saps, M., Sudakaran, S., Deb, C., & Mehta, D. (2022). Gut Microbiome Remains Static in Functional Abdominal Pain Disorders Patients Compared to Controls: Potential for Diagnostic Tools. BioTech, 11(4), 50. https://doi.org/10.3390/biotech11040050






