Cardiovascular Diseases in the Digital Health Era: A Translational Approach from the Lab to the Clinic
Abstract
1. Introduction
2. A Translational Approach in Cardiovascular Diseases: Chimera or Reality?
2.1. The Present Breach among Basic Biomedical Research and Clinical Applications
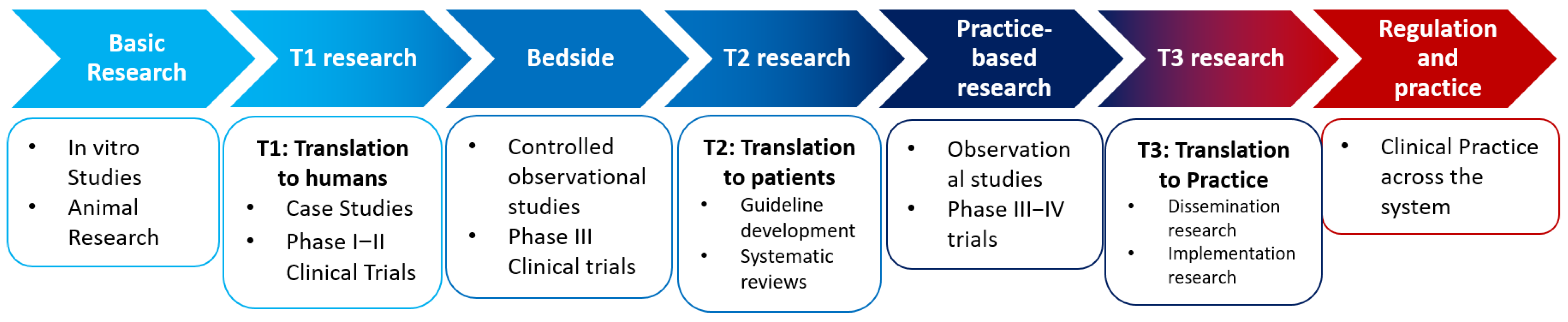
2.2. Translational Research as a Highly Complex Structured Matrix
- To establish better preclinical models that allow researchers to rationally select target compounds and to better understand their mechanism of action.
- To evaluate and incorporate clear endpoints at preclinical stages that allow for anoptimal evaluation of target-based new drugs.
- To define current monitoring techniques that help to develop the tools, probes, and biological and imaging assays suitable for in vitro assessment, in preclinical models.
- To conduct, in a rapid, coordinated manner, highly specialized, complex, early clinical trials with rigorous standards to deliver complex, detailed data for licensing purposes.
- To ensure a high-quality laboratory infrastructure and expertise with the capacity to provide biological readouts on clinical material in a timely manner.
2.3. Current Accomplishments in Cardiovascular Health
2.3.1. Translational Bioinformatics
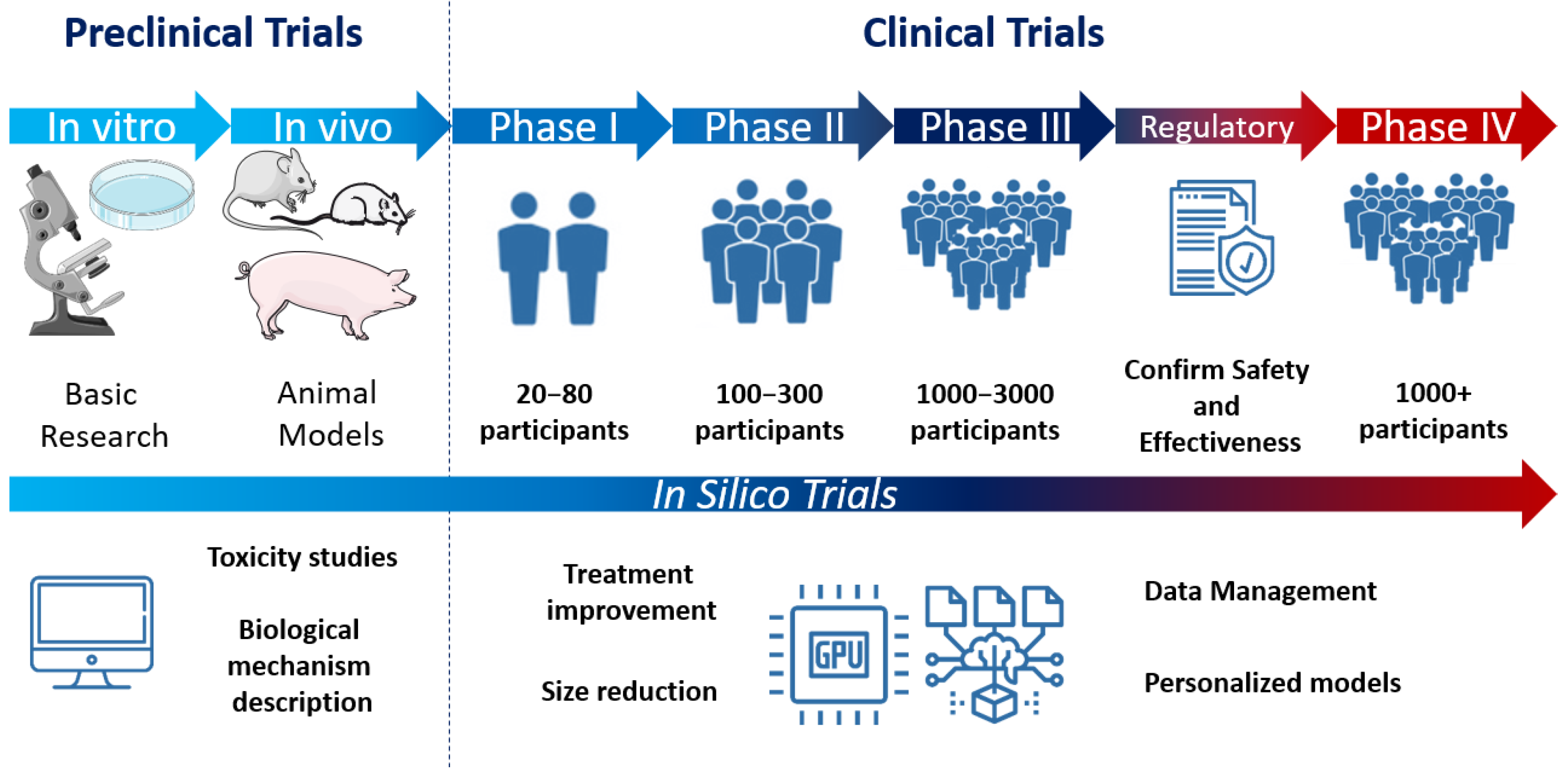
2.3.2. Computational Models for Personalized Medicine
- (1)
- Reducing the size or studying specific groups at the clinical level that are identified as risk groups at in silico level.
- (2)
- Adding more detailed information obtained from this type of trials to better understand interactions with different groups and long-term effects that clinical trials cannot provide.
- (3)
- Replacing the preclinical phase and preserving the clinical trial for legal requirements.
- (4)
- Improving unsuccessful treatments or products by providing extra information, as this increases innovation, decreases economical costs, and exponentially increases the understanding of biological processes.
- (5)
- Avoiding the use of animal models by directly including clinical data and personalized information from the patients. This significantly decreases the overall costs associated with the development of treatments and has proven to be more effective at predicting the behavior of the drug or treatment in large-scale trials and identifying secondary effects, therefore better screening the treatments that progress to phase III clinical trials.
2.3.3. In Vitro Research and Translational In Vitro Diagnostics
- (1)
- Inappropriate patient sample or signal acquisition that leads to an inability to analyze the data.
- (2)
- Difficulties or deterioration of the sample during its collection, management, treatment, storage, or transport, especially for biological samples.
- (3)
- Inability to afford in vitro testing at large scales or highly efficient computational systems that can analyze large amounts of data.
2.3.4. Animal Models as a Translational Model for Research
2.3.5. Signal Acquisition and Processing Automation Using Artificial Intelligence
2.4. Economical Issues and Legal Regulations
- (1)
- Regulatory authorities’ actions against digital health and healthcare IT that meet the definition of medical devices but have not obtained the CE mark.
- (2)
- The European Data Protection Agency’s actions in the event of breaches of data protection legislation and data security.
3. Current Trends and Future Perspectives
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Saner, H.; Van Der Velde, E. eHealth in Cardiovascular Medicine: A Clinical Update. Eur. J. Prev. Cardiol. 2016, 23, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Pea, A.; Scarpa, A. Artificial Intelligence in Oncology: Current Applications and Future Perspectives. Br. J. Cancer 2021, 126, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.; Verspoor, K.; Jenkinson, M.; Law, M.; Abbott, D.F.; Jackson, G.D. Artificial Intelligence for Clinical Decision Support in Neurology. Brain Commun. 2020, 2, fcaa096. [Google Scholar] [CrossRef] [PubMed]
- De Marvao, A.; Dawes, T.J.W.; Howard, J.P.; O’Regan, D.P. Artificial Intelligence and the Cardiologist: What You Need to Know for 2020. Heart 2020, 106, 399–400. [Google Scholar] [CrossRef] [PubMed]
- Visseren, F.L.J.; MacH, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Bäck, M.; Benetos, A.; Biffi, A.; Boavida, J.M.; Capodanno, D.; et al. 2021 ESC Guidelines on Cardiovascular Disease Prevention in Clinical Practice. Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef] [PubMed]
- WHO|Cardiovascular Diseases (CVDs). WHO 2016. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 21 April 2022).
- Cardiovascular Diseases—World Health Organization. Available online: https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_3 (accessed on 21 April 2022).
- Morris, Z.S.; Wooding, S.; Grant, J. The Answer Is 17 Years, What Is the Question: Understanding Time Lags in Translational Research. J. R. Soc. Med. 2011, 104, 510. [Google Scholar] [CrossRef]
- Westfall, J.M.; Mold, J.; Fagnan, L. Practice-Based Research—“Blue Highways” on the NIH Roadmap. JAMA 2007, 297, 403–406. [Google Scholar] [CrossRef]
- Rubio, D.M.G.; Schoenbaum, E.E.; Lee, L.S.; Schteingart, D.E.; Marantz, P.R.; Anderson, K.E.; Platt, L.D.; Baez, A.; Esposito, K. Defining Translational Research: Implications for Training. Acad. Med. 2010, 85, 470. [Google Scholar] [CrossRef]
- Wolf, S.M.; Clayton, E.W.; Lawrenz, F. Introduction: The Crucial Role of Law in Supporting Successful Translation of Genomics into Clinical Care. J. Law Med. Ethics 2020, 48, 7–10. [Google Scholar] [CrossRef]
- McMahon, G.T.; Katz, J.T.; Thorndike, M.E.; Levy, B.D.; Loscalzo, J. Evaluation of a Redesign Initiative in an Internal-Medicine Residency. N. Engl. J. Med. 2010, 362, 1304–1311. [Google Scholar] [CrossRef]
- Austin, C.P. Opportunities and Challenges in Translational Science. Clin. Transl. Sci. 2021, 14, 1629–1647. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, F.; Lacombe, D.; Therasse, P.; Eggermont, A.M.M. Integration of Translational Research in the European Organization for Research and Treatment of Cancer Research (EORTC) Clinical Trial Cooperative Group Mechanisms. J. Transl. Med. 2003, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Avilés, F.; Sanz-Ruiz, R.; Climent, A.M.; Badimon, L.; Bolli, R.; Charron, D.; Fuster, V.; Janssens, S.; Kastrup, J.; Kim, H.S.; et al. Global Position Paper on Cardiovascular Regenerative Medicine. Eur. Heart J. 2017, 38, 2532–2546. [Google Scholar] [CrossRef] [PubMed]
- Svennberg, E.; Tjong, F.; Goette, A.; Akoum, N.; Di Biaise, L.; Bordachar, P.; Boriani, G.; Burri, H.; Conte, G.; Deharo, J.-C.; et al. How to Use Digital Devices to Detect and Manage Arrhythmias: An EHRA Practical Guide. EP Eur. 2022, euac038. [Google Scholar] [CrossRef]
- Qazi, S.; Raza, K. Translational Bioinformatics in Healthcare: Past, Present, and Future. Transl. Bioinform. Healthc. Med. 2021, 13, 1–12. [Google Scholar] [CrossRef]
- Khomtchouk, B.B.; Tran, D.T.; Vand, K.A.; Might, M.; Gozani, O.; Assimes, T.L. Cardioinformatics: The Nexus of Bioinformatics and Precision Cardiology. Brief. Bioinform. 2020, 21, 2031–2051. [Google Scholar] [CrossRef]
- Houser, S.R. The American Heart Association’s New Institute for Precision Cardiovascular Medicine. Circulation 2016, 134, 1913–1914. [Google Scholar] [CrossRef]
- Mao, C.; Howard, T.D.; Sullivan, D.; Fu, Z.; Yu, G.; Parker, S.J.; Will, R.; Vander Heide, R.S.; Wang, Y.; Hixson, J.; et al. Bioinformatic Analysis of Coronary Disease Associated SNPs and Genesto Identify Proteins Potentially Involved in the Pathogenesis of atherosclerosis. J. Proteom. Genom. Res. 2017, 2, 1. [Google Scholar] [CrossRef]
- O’Leary, K. AI Refines Treatment Selection for Heart Failure. Nat. Med. 2021. [Google Scholar] [CrossRef]
- Fernández, A.I.; Bermejo, J.; Yotti, R.; Martínez-Gonzalez, M.Á.; Mira, A.; Gophna, U.; Karlsson, R.; Al-Daccak, R.; Martín-Demiguel, I.; Gutiérrez-Ibanes, E.; et al. The Impact of Mediterranean Diet on Coronary Plaque Vulnerability, Microvascular Function, Inflammation and Microbiome after an Acute Coronary Syndrome: Study Protocol for the MEDIMACS Randomized, Controlled, Mechanistic Clinical Trial. Trials 2021, 22, 795. [Google Scholar] [CrossRef]
- Li, Y.; Meng, H.; Liu, Y.; Lee, B.P. Fibrin Gel as an Injectable Biodegradable Scaffold and Cell Carrier for Tissue Engineering. Sci. World J. 2015, 2015, 685690. [Google Scholar] [CrossRef] [PubMed]
- Passini, E.; Britton, O.J.; Lu, H.R.; Rohrbacher, J.; Hermans, A.N.; Gallacher, D.J.; Greig, R.J.H.; Bueno-Orovio, A.; Rodriguez, B. Human In Silico Drug Trials Demonstrate Higher Accuracy than Animal Models in Predicting Clinical Pro-Arrhythmic Cardiotoxicity. Front. Physiol. 2017, 8, 668. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Stohlman, J.; Dang, Q.; Strauss, D.G.; Blinova, K. Assessment of Proarrhythmic Potential of Drugs in Optogenetically Paced Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Toxicol. Sci. 2019, 170, 167–179. [Google Scholar] [CrossRef]
- Vicente, J.; Zusterzeel, R.; Johannesen, L.; Ochoa-Jimenez, R.; Mason, J.W.; Sanabria, C.; Kemp, S.; Sager, P.T.; Patel, V.; Matta, M.K.; et al. Assessment of Multi-Ion Channel Block in a Phase I Randomized Study Design: Results of the CiPA Phase I ECG Biomarker Validation Study. Clin. Pharmacol. Ther. 2019, 105, 943–953. [Google Scholar] [CrossRef]
- Liberos, A.; Bueno-Orovio, A.; Rodrigo, M.; Ravens, U.; Hernandez-Romero, I.; Fernandez-Aviles, F.; Guillem, M.S.; Rodriguez, B.; Climent, A.M. Balance between Sodium and Calcium Currents Underlying Chronic Atrial Fibrillation Termination: An in Silico Intersubject Variability Study. Hear. Rhythm 2016, 13, 2358–2365. [Google Scholar] [CrossRef]
- Vigmond, E.; Vadakkumpadan, F.; Gurev, V.; Arevalo, H.; Deo, M.; Plank, G.; Trayanova, N. Towards Predictive Modelling of the Electrophysiology of the Heart. Exp. Physiol. 2009, 94, 563–577. [Google Scholar] [CrossRef]
- Arevalo, H.J.; Vadakkumpadan, F.; Guallar, E.; Jebb, A.; Malamas, P.; Wu, K.C.; Trayanova, N.A. Arrhythmia Risk Stratification of Patients after Myocardial Infarction Using Personalized Heart Models. Nat. Commun. 2016, 7, 11437. [Google Scholar] [CrossRef]
- Rivera-Juárez, A.; Hernández-Romero, I.; Puertas, C.; Zhang-Wang, S.; Sánchez-Álamo, B.; Martins, R.; Figuera, C.; Guillem, M.S.; Climent, A.M.; Fernández-Avilés, F.; et al. Clinical Characteristics and Electrophysiological Mechanisms Underlying Brugada ECG in Patients With Severe Hyperkalemia. J. Am. Hear. Assoc. Cardiovasc. Cerebrovasc. Dis. 2019, 8, e010115. [Google Scholar] [CrossRef]
- Carro, J.; Rodríguez-Matas, J.F.; Monasterio, V.; Pueyo, E. Limitations in Electrophysiological Model Development and Validation Caused by Differences between Simulations and Experimental Protocols. Prog. Biophys. Mol. Biol. 2017, 129, 53–64. [Google Scholar] [CrossRef]
- Crumb, W.J.; Vicente, J.; Johannesen, L.; Strauss, D.G. An Evaluation of 30 Clinical Drugs against the Comprehensive in Vitro Proarrhythmia Assay (CiPA) Proposed Ion Channel Panel. J. Pharmacol. Toxicol. Methods 2016, 81, 251–262. [Google Scholar] [CrossRef]
- de la Nava, A.M.S.; Mansilla, A.G.; González-Torrecilla, E.; Ávila, P.; Datino, T.; Bermejo, J.; Arenal, Á.; Fernández-Avilés, F.; Atienza, F. Personalized Evaluation of Atrial Complexity of Patients Undergoing Atrial Fibrillation Ablation: A Clinical Computational Study. Biology 2021, 10, 838. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Simundic, A.M.; Rodriguez-Manas, L.; Bossuyt, P.; Banfi, G. Standardizing in Vitro Diagnostics Tasks in Clinical Trials: A Call for Action. Ann. Transl. Med. 2016, 4, 181. [Google Scholar] [CrossRef] [PubMed]
- Montero, P.; Flandes-Iparraguirre, M.; Musquiz, S.; Pérez Araluce, M.; Plano, D.; Sanmartín, C.; Orive, G.; Gavira, J.J.; Prosper, F.; Mazo, M.M. Cells, Materials, and Fabrication Processes for Cardiac Tissue Engineering. Front. Bioeng. Biotechnol. 2020, 8, 955. [Google Scholar] [CrossRef]
- Gómez-Cid, L.; López-Donaire, M.L.; Velasco, D.; Marín, V.; González, M.I.; Salinas, B.; Cussó, L.; García, Á.; Bravo, S.B.; Fernández-Santos, M.E.; et al. Cardiac Extracellular Matrix Hydrogel Enriched with Polyethylene Glycol Presents Improved Gelation Time and Increased On-Target Site Retention of Extracellular Vesicles. Int. J. Mol. Sci. 2021, 22, 9226. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, P.L.; Fernández-Santos, M.E.; Costanza, S.; Climent, A.M.; Moscoso, I.; Gonzalez-Nicolas, M.A.; Sanz-Ruiz, R.; Rodríguez, H.; Kren, S.M.; Garrido, G.; et al. Acellular Human Heart Matrix: A Critical Step toward Whole Heart Grafts. Biomaterials 2015, 61, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Haake, K.; Kempf, H.; Kaschutnig, P.; Weiss, A.C.; Nguyen, A.H.H.; Abeln, M.; Merkert, S.; Kühnel, M.P.; Hartmann, D.; et al. A 3D IPSC-Differentiation Model Identifies Interleukin-3 as a Regulator of Early Human Hematopoietic Specification. Haematologica 2021, 106, 1354–1367. [Google Scholar] [CrossRef]
- Lewandowski, J.; Rozwadowska, N.; Kolanowski, T.J.; Malcher, A.; Zimna, A.; Rugowska, A.; Fiedorowicz, K.; Łabędź, W.; Kubaszewski, Ł.; Chojnacka, K.; et al. The Impact of in Vitro Cell Culture Duration on the Maturation of Human Derived from Induced Pluripotent Stem Cells of Myogenic. Cell Transplant. 2018, 27, 1047. [Google Scholar] [CrossRef]
- Yang, X.; Rodriguez, M.L.; Leonard, A.; Sun, L.; Fischer, K.A.; Wang, Y.; Ritterhoff, J.; Zhao, L.; Kolwicz, S.C.; Pabon, L.; et al. Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2019, 13, 657. [Google Scholar] [CrossRef]
- Vuorenpää, H.; Penttinen, K.; Heinonen, T.; Pekkanen-Mattila, M.; Sarkanen, J.R.; Ylikomi, T.; Aalto-Setälä, K. Maturation of Human Pluripotent Stem Cell Derived Cardiomyocytes Is Improved in Cardiovascular Construct. Cytotechnology 2017, 69, 785. [Google Scholar] [CrossRef]
- Kroll, K.; Chabria, M.; Wang, K.; Häusermann, F.; Schuler, F.; Polonchuk, L. Electro-Mechanical Conditioning of Human IPSC-Derived Cardiomyocytes for Translational Research. Prog. Biophys. Mol. Biol. 2017, 130, 212–222. [Google Scholar] [CrossRef]
- Cho, G.S.; Lee, D.I.; Tampakakis, E.; Murphy, S.; Andersen, P.; Uosaki, H.; Chelko, S.; Chakir, K.; Hong, I.; Seo, K.; et al. Neonatal Transplantation Confers Maturation of PSC-Derived Cardiomyocytes Conducive to Modeling Cardiomyopathy. Cell Rep. 2017, 18, 571. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.K.; Gupta, R. Artificial Intelligence Methods for Analysis of Electrocardiogram Signals for Cardiac Abnormalities: State-of-the-Art and Future Challenges. Artif. Intell. Rev. 2021, 55, 1519–1565. [Google Scholar] [CrossRef]
- Grankvist, R.; Chireh, A.; Sandell, M.; Mukarram, A.K.; Jaff, N.; Berggren, I.; Persson, H.; Linde, C.; Arnberg, F.; Lundberg, J.; et al. Myocardial Micro-Biopsy Procedure for Molecular Characterization with Increased Precision and Reduced Trauma. Sci. Rep. 2020, 10, 8029. [Google Scholar] [CrossRef] [PubMed]
- Scholar, R.; Singaraju, J. Decision Support System for Congenital Heart Disease Diagnosis Based on Signs and Symptoms Using Neural Networks Vanisree K. Int. J. Comput. Appl. 2011, 19, 975–8887. [Google Scholar]
- Moore, R.A.; Madueme, P.C.; Lorts, A.; Morales, D.L.S.; Taylor, M.D. Virtual Implantation Evaluation of the Total Artificial Heart and Compatibility: Beyond Standard Fit Criteria. J. Hear. Lung Transpl. 2014, 33, 1180–1183. [Google Scholar] [CrossRef]
- Abudan, A.A.; Isath, A.; Ryan, J.D.; Henrich, M.J.; Dugan, J.L.; Attia, Z.I.; Ladewig, D.J.; Dillon, J.J.; Friedman, P.A. Safety and Compatibility of Smart Device Heart Rhythm Monitoring in Patients with Cardiovascular Implantable Electronic Devices. J. Cardiovasc. Electrophysiol. 2019, 30, 1602–1609. [Google Scholar] [CrossRef]
- Vargas, J.E. Home-Based Monitoring of Cardiac Patients. In Proceedings of the 1998 IEEE International Conference on Information Technology Applications in Biomedicine, ITAB 1998, Washington, DC, USA, 17 May 1998; pp. 133–136. [Google Scholar] [CrossRef]
- Denayer, T.; Stöhrn, T.; Van Roy, M. Animal Models in Translational Medicine: Validation and Prediction. New Horizons Transl. Med. 2014, 2, 5–11. [Google Scholar] [CrossRef]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is It Time to Rethink Our Current Approach? JACC Basic Transl. Sci. 2019, 4, 845–854. [Google Scholar] [CrossRef]
- Lawrence, C.L.; Bridgland-Taylor, M.H.; Pollard, C.E.; Hammond, T.G.; Valentin, J.-P. A Rabbit Langendorff Heart Proarrhythmia Model: Predictive Value for Clinical Identification of Torsades de Pointes. Br. J. Pharmacol. 2006, 149, 845–860. [Google Scholar] [CrossRef]
- von Kortzfleisch, V.T.; Karp, N.A.; Palme, R.; Kaiser, S.; Sachser, N.; Richter, S.H. Improving Reproducibility in Animal Research by Splitting the Study Population into Several ‘Mini-Experiments’. Sci. Rep. 2020, 10, 16579. [Google Scholar] [CrossRef]
- Wu, Y.; Yin, X.; Wijaya, C.; Huang, M.H.; McConnell, B.K. Acute Myocardial Infarction in Rats. J. Vis. Exp. 2011, 48, e2464. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, M.L.; Brunt, K.R.; Kirk, J.A.; Kleinbongard, P.; Calvert, J.W.; de Castro Brás, L.E.; DeLeon-Pennell, K.Y.; Del Re, D.P.; Frangogiannis, N.G.; Frantz, S.; et al. Guidelines for in Vivo Mouse Models of Myocardial Infarction. Am. J. Physiol. Hear. Circ. Physiol. 2021, 321, H1056–H1073. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Morimoto, Y.; Ishihara, M.; Shimizu, M.; Takase, B.; Maehara, T.; Kikuchi, M. A New Rabbit Model of Myocardial Infarction without Endotracheal Intubation. J. Surg. Res. 2004, 116, 124–128. [Google Scholar] [CrossRef]
- Polizzotti, B.D.; Ganapathy, B.; Haubner, B.J.; Penninger, J.M.; Kühn, B. A Cryoinjury Model in Neonatal Mice for Cardiac Translational and Regeneration Research. Nat. Protoc. 2016, 11, 542. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, R.; Cao, G.; Zhang, F.; Zhang, Y.; Zhang, Z.; Wu, S. A Reliable Rabbit Model for Hyperkinetic Pulmonary Hypertension. J. Thorac. Cardiovasc. Surg. 2010, 140, 395–399. [Google Scholar] [CrossRef][Green Version]
- Odening, K.E.; Baczko, I.; Brunner, M.; Mechanisms, K.G.; de Medeiros, R.A.; Hausen, Z.A. Animals in Cardiovascular Research: Important Role of Rabbit Models in Cardiac Electrophysiology. Eur. Heart J. 2020, 41, 2036. [Google Scholar] [CrossRef]
- Powers, J.C.; Recchia, F. Canine Model of Pacing-Induced Heart Failure. Methods Mol. Biol. 2018, 1816, 309–325. [Google Scholar] [CrossRef]
- Crisóstomo, V.; Sun, F.; Maynar, M.; Báez-Díaz, C.; Blanco, V.; Garcia-Lindo, M.; Usón-Gargallo, J.; Sánchez-Margallo, F.M. Common Swine Models of Cardiovascular Disease for Research and Training. Lab Anim. 2016, 45, 67–74. [Google Scholar] [CrossRef]
- Tsang, H.G.; Rashdan, N.A.; Whitelaw, C.B.A.; Corcoran, B.M.; Summers, K.M.; MacRae, V.E. Large Animal Models of Cardiovascular Disease. Cell Biochem. Funct. 2016, 34, 113. [Google Scholar] [CrossRef]
- Chamuleau, S.A.J.; Van Der Naald, M.; Climent, A.M.; Kraaijeveld, A.O.; Wever, K.E.; Duncker, D.J.; Fernández-Avilés, F.; Bolli, R. Translational Research in Cardiovascular Repair a Call for a Paradigm Shift. Circ. Res. 2018, 122, 310–318. [Google Scholar] [CrossRef]
- Povsic, T.J.; Sanz-Ruiz, R.; Climent, A.M.; Bolli, R.; Taylor, D.A.; Gersh, B.J.; Menasché, P.; Perin, E.C.; Pompilio, G.; Atsma, D.E.; et al. Reparative Cell Therapy for the Heart: Critical Internal Appraisal of the Field in Response to Recent Controversies. ESC Hear. Fail. 2021, 8, 2306–2309. [Google Scholar] [CrossRef] [PubMed]
- Grigorian-Shamagian, L.; Sanz-Ruiz, R.; Climent, A.; Badimon, L.; Barile, L.; Bolli, R.; Chamuleau, S.; Grobbee, D.E.; Janssens, S.; Kastrup, J.; et al. Insights into Therapeutic Products, Preclinical Research Models, and Clinical Trials in Cardiac Regenerative and Reparative Medicine: Where Are We Now and the Way Ahead. Current Opinion Paper of the ESC Working Group on Cardiovascular Regenerative and Reparative Medicine. Cardiovasc. Res. 2021, 117, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Madonna, R.; Van Laake, L.W.; Davidson, S.M.; Engel, F.B.; Hausenloy, D.J.; Lecour, S.; Leor, J.; Perrino, C.; Schulz, R.; Ytrehus, K.; et al. Position Paper of the European Society of Cardiology Working Group Cellular Biology of the Heart: Cell-Based Therapies for Myocardial Repair and Regeneration in Ischemic Heart Disease and Heart Failure. Eur. Heart J. 2016, 37, 1789–1798. [Google Scholar] [CrossRef] [PubMed]
- Grigorian Shamagian, L.; Madonna, R.; Taylor, D.; Climent, A.M.; Prosper, F.; Bras-Rosario, L.; Bayes-Genis, A.; Ferdinandy, P.; Fernández-Avilés, F.; Izpisua Belmonte, J.C.; et al. Perspectives on Directions and Priorities for Future Preclinical Studies in Regenerative Medicine. Circ. Res. 2019, 124, 938–951. [Google Scholar] [CrossRef]
- Allen, B.; Seltzer, S.E.; Langlotz, C.P.; Dreyer, K.P.; Summers, R.M.; Petrick, N.; Marinac-Dabic, D.; Cruz, M.; Alkasab, T.K.; Hanisch, R.J.; et al. A Road Map for Translational Research on Artificial Intelligence in Medical Imaging: From the 2018 National Institutes of Health/RSNA/ACR/The Academy Workshop. J. Am. Coll. Radiol. 2019, 16, 1179–1189. [Google Scholar] [CrossRef]
- Sanchez de la Nava, A.M.; Arenal, Á.; Fernández-Avilés, F.; Atienza, F. Artificial Intelligence-Driven Algorithm for Drug Effect Prediction on Atrial Fibrillation: An in Silico Population of Models Approach. Front. Physiol. 2021, 2079. [Google Scholar] [CrossRef]
- Sánchez de la Nava, A.M.; Atienza, F.; Bermejo, J.; Fernández-Avilés, F. Artificial Intelligence for a Personalized Diagnosis and Treatment of Atrial Fibrillation. Am. J. Physiol. Circ. Physiol. 2021, 320, H1337–H1347. [Google Scholar] [CrossRef]
- Ríos-Muñoz, G.R.; Fernández-Avilés, F.; Arenal, Á. Convolutional Neural Networks for Mechanistic Driver Detection in Atrial Fibrillation. Int. J. Mol. Sci. 2022, 23, 4216. [Google Scholar] [CrossRef]
- Deo, R.C. Machine Learning in Medicine: Will This Time Be Different? Circulation 2020, 142, 1521–1523. [Google Scholar] [CrossRef]
- Kim, D.W.; Jang, H.Y.; Kim, K.W.; Shin, Y.; Park, S.H. Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean J. Radiol. 2019, 20, 405–410. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, J.W.; Zang, G.Y.; Pu, J. The Primary Use of Artificial Intelligence in Cardiovascular Diseases: What Kind of Potential Role Does Artificial Intelligence Play in Future Medicine? J. Geriatr. Cardiol. 2019, 16, 585. [Google Scholar] [CrossRef] [PubMed]
- Cowie, M.R.; Lam, C.S.P. Remote Monitoring and Digital Health Tools in CVD Management. Nat. Rev. Cardiol. 2021, 18, 457–458. [Google Scholar] [CrossRef]
- Sharma, A.; Zheng, Y.; Ezekowitz, J.A.; Westerhout, C.M.; Udell, J.A.; Goodman, S.G.; Armstrong, P.W.; Buse, J.B.; Green, J.B.; Josse, R.G.; et al. Cluster Analysis of Cardiovascular Phenotypes in Patients With Type 2 Diabetes and Established Atherosclerotic Cardiovascular Disease: A Potential Approach to Precision Medicine. Diabetes Care 2022, 45, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Jahangiry, L.; Abbasalizad Farhangi, M.; Najafi, M.; Sarbakhsh, P. Clusters of the Risk Markers and the Pattern of Premature Coronary Heart Disease: An Application of the Latent Class Analysis. Front. Cardiovasc. Med. 2021, 8, 1744. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Bin, G.; Wu, S.; Bin, G.; Huang, J.; Zhou, Z. Detection of Atrial Fibrillation from ECG Recordings Using Decision Tree Ensemble with Multi-Level Features. Physiol. Meas. 2018, 39, 094008. [Google Scholar] [CrossRef] [PubMed]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia Detection Using Deep Convolutional Neural Network with Long Duration ECG Signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Krittanawong, C.; Virk, H.U.H.; Bangalore, S.; Wang, Z.; Johnson, K.W.; Pinotti, R.; Zhang, H.J.; Kaplin, S.; Narasimhan, B.; Kitai, T.; et al. Machine Learning Prediction in Cardiovascular Diseases: A Meta-Analysis. Sci. Rep. 2020, 10, 16057. [Google Scholar] [CrossRef]
- High-Level Expert Group on Artificial Intelligence Set Up by the European Commission Ethics Guidelines for Trustworthy AI. Available online: https://ec.europa.eu/futurium/en/ai-alliance-consultation/guidelines.1.html (accessed on 22 April 2022).
- Six Factors Affecting Reproducibility in Life Science Research and How to Handle Them. Available online: https://www.nature.com/articles/d42473-019-00004-y (accessed on 22 April 2022).
- Begley, C.G.; Ioannidis, J.P.A. Reproducibility in Science: Improving the Standard for Basic and Preclinical Research. Circ. Res. 2015, 116, 116–126. [Google Scholar] [CrossRef]
- Meslin, E.M.; Blasimme, A.; Cambon-Thomsen, A. Mapping the Translational Science Policy “Valley of Death”. Clin. Transl. Med. 2013, 2, 1. [Google Scholar] [CrossRef]
- Pearson, A.; Jordan, Z.; Munn, Z. Translational Science and Evidence-Based Healthcare: A Clarification and Reconceptualization of How Knowledge Is Generated and Used in Healthcare. Nurs. Res. Pract. 2012, 2012, 792519. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez de la Nava, A.M.; Gómez-Cid, L.; Ríos-Muñoz, G.R.; Fernández-Santos, M.E.; Fernández, A.I.; Arenal, Á.; Sanz-Ruiz, R.; Grigorian-Shamagian, L.; Atienza, F.; Fernández-Avilés, F. Cardiovascular Diseases in the Digital Health Era: A Translational Approach from the Lab to the Clinic. BioTech 2022, 11, 23. https://doi.org/10.3390/biotech11030023
Sánchez de la Nava AM, Gómez-Cid L, Ríos-Muñoz GR, Fernández-Santos ME, Fernández AI, Arenal Á, Sanz-Ruiz R, Grigorian-Shamagian L, Atienza F, Fernández-Avilés F. Cardiovascular Diseases in the Digital Health Era: A Translational Approach from the Lab to the Clinic. BioTech. 2022; 11(3):23. https://doi.org/10.3390/biotech11030023
Chicago/Turabian StyleSánchez de la Nava, Ana María, Lidia Gómez-Cid, Gonzalo Ricardo Ríos-Muñoz, María Eugenia Fernández-Santos, Ana I. Fernández, Ángel Arenal, Ricardo Sanz-Ruiz, Lilian Grigorian-Shamagian, Felipe Atienza, and Francisco Fernández-Avilés. 2022. "Cardiovascular Diseases in the Digital Health Era: A Translational Approach from the Lab to the Clinic" BioTech 11, no. 3: 23. https://doi.org/10.3390/biotech11030023
APA StyleSánchez de la Nava, A. M., Gómez-Cid, L., Ríos-Muñoz, G. R., Fernández-Santos, M. E., Fernández, A. I., Arenal, Á., Sanz-Ruiz, R., Grigorian-Shamagian, L., Atienza, F., & Fernández-Avilés, F. (2022). Cardiovascular Diseases in the Digital Health Era: A Translational Approach from the Lab to the Clinic. BioTech, 11(3), 23. https://doi.org/10.3390/biotech11030023






