Increased Detection of Merkel Cell Polyomavirus in Non-Melanoma Skin Cancer and Its Association with Host Immunogenetic Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Molecular Diagnosis of MCPyV and Cytokine Genotyping
2.3. Statistical Analysis
3. Results
3.1. MCPyV Detection by Age, Sex, Ethnicity, and Lesion Type
3.2. Variability of MCPyV Detection According to Tissue Source and Sun Exposure
3.3. Association Between MCPyV Detection and Cytokine Gene Polymorphisms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MCPyV | Merkel cell polyomavirus |
| NMSC | Non-melanoma skin cancer |
| qPCR | Real-time PCR |
| BCC | Basal cell carcinoma |
| SCC | Squamous cell carcinoma |
| MCC | Merkel cell carcinoma |
| IL-6 | Interleukin-6 |
| IL-10 | Interleukin-10 |
| TNF-α | Tumor necrosis factor-alpha |
| IFN-γ | Interferon-gamma |
References
- Hu, W.; Fang, L.; Ni, R.; Zhang, H.; Pan, G. Changing trends in the disease burden of non-melanoma skin cancer globally from 1990 to 2019 and its predicted level in 25 years. J. Med. Virol. 2022, 22, 836. [Google Scholar] [CrossRef] [PubMed]
- IARC. Skin cancer. World Health Organization. J. Med. Virol. 2020.
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Bellott, T.R.; Luz, F.B.; Silva, A.K.F.D.; Varella, R.B.; Rochael, M.C.; Pantaleão, L. Merkel cell polyomavirus and its etiological relationship with skin carcinomas. An. Bras. Dermatol. 2023, 96, 332–337. [Google Scholar]
- Tribble, J.T.; Pfeiffer, R.M.; Brownell, I.; Cahoon, E.K.; Sargen, M.R.; Shiels, M.S.; Luo, Q.; Cohen, C.; Drezner, K.; Hernandez, B.; et al. Merkel cell carcinoma and immunosuppression, UV radiation, and Merkel cell polyomavirus. JAMA Dermatol. 2025, 161, 47–55. [Google Scholar] [CrossRef]
- Rollison, D.E.; Giuliano, A.R.; Becker, J.C. New virus associated with Merkel cell carcinoma development. J. Natl. Compr. Cancer Netw. 2010, 8, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Krump, N.A.; You, J. From Merkel cell polyomavirus infection to Merkel cell carcinoma oncogenesis. Front. Microbiol. 2021, 12, 739695. [Google Scholar] [CrossRef]
- Becker, J.C.; Stang, A.; DeCaprio, J.A.; Cerroni, L.; Lebbé, C.; Veness, M.; Nghiem, P. Merkel cell carcinoma. Nat. Rev. Dis. Primers 2017, 3, 17077. [Google Scholar] [CrossRef]
- Bhatia, K.; Goedert, J.J.; Modali, R.; Preiss, L.; Ayers, L.W. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. Int. J. Cancer 2010, 126, 2240–2246. [Google Scholar] [CrossRef]
- Kassem, A.; Schöpflin, A.; Diaz, C.; Weyers, W.; Stickeler, E.; Werner, M.; Zur Hausen, A. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008, 68, 5009–5013. [Google Scholar] [CrossRef] [PubMed]
- Houben, R.; Shuda, M.; Weinkam, R.; Schrama, D.; Feng, H.; Chang, Y.; Moore, P.S.; Becker, J.C. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010, 84, 7064–7072. [Google Scholar] [CrossRef]
- Moreno, M.; Schmitt, R.L.; Lang, M.G.; Gheno, V. Epidemiological profile of patients with cutaneous melanoma in a region of southern Brazil. J. Ski. Cancer 2012, 2012, 917346. [Google Scholar] [CrossRef] [PubMed]
- Shuda, M.; Arora, R.; Kwun, H.J.; Feng, H.; Sarid, R.; Fernández-Figueras, M.T.; Tolstov, Y.; Gjoerup, O.; Mansukhani, M.M.; Swerdlow, S.H.; et al. Human Merkel cell polyomavirus infection. II. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int. J. Cancer 2009, 125, 1250–1256. [Google Scholar] [CrossRef]
- Al Kadi, M.; Monem, F. Polymorphism of IFN-γ (+874 T/A) in Syrian patients with chronic hepatitis B. Gastroenterol. Hepatol. Bed Bench 2017, 10, 34–38. [Google Scholar] [PubMed]
- Joshi, L.; Ponnana, M.; Sivangala, R.; Chelluri, L.K.; Nallari, P.; Penmetsa, S.; Valluri, V.; Gaddam, S. Evaluation of TNF-α, IL-10 and IL-6 cytokine production and their correlation with genotype variants amongst tuberculosis patients and their household contacts. PLoS ONE 2015, 10, e0137727. [Google Scholar] [CrossRef]
- da Silva, N.M.O.; Germano, F.N.; Vidales-Braz, B.M.; do Carmo Zanella, R.; dos Santos, D.M.; Lobato, R.; de Martinez, A.M.B. Polymorphisms of IL-10 gene in patients infected with HCV under antiviral treatment in southern Brazil. Cytokine 2015, 73, 253–257. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Taghizadieh, M.; Baradaran, B.; Yousefi, M.; Shokri, M.; Hajiasgharzadeh, K.; Estiar, M.A. Association between Merkel cell polyomavirus (MCPyV) large T antigen load and nonmelanoma skin cancers. Pathol. Res. Pract. 2022, 238, 154112. [Google Scholar]
- Tuttleton Arron, S.; Jennings, L.; Nindl, I.; Rosl, F.; Bouwes Bavinck, J.N.; Seçkin, D.; Trakatelli, M.; Murphy, G.M.; Viral Working Group of the International Transplant Skin Cancer Collaborative (ITSCC); Skin Care in Organ Transplant Patients, Europe (SCOPE). Viral oncogenesis and its role in nonmelanoma skin cancer. Br. J. Dermatol. 2011, 164, 1201–1213. [Google Scholar] [CrossRef]
- Baez, C.F.; Gonçalves, M.T.V.; da Rocha, W.M.; Magalhães de Souza, L.; Savassi-Ribas, F.; de Oliveira Almeida, N.K.; Delbue, S.; Guimarães, M.A.A.M.; Cavalcanti, S.M.B.; Luz, F.B.; et al. Investigation of three oncogenic epitheliotropic viruses shows human papillomavirus in association with non-melanoma skin cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, T. The dark and the sunny sides of UVR-induced immunosuppression: Photoimmunology revisited. J. Investig. Dermatol. 2010, 130, 49–54. [Google Scholar] [CrossRef]
- Redpath, S.; Ghazal, P.; Gascoigne, N.R.J. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001, 9, 86–92. [Google Scholar] [CrossRef]
- Brooks, D.G.; Trifilo, M.J.; Edelmann, K.H.; Teyton, L.; McGavern, D.B.; Oldstone, M.B. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 2006, 12, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Karin, M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 2018, 35, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The role of interleukin 6 during viral infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef]
- Wong, S.Q.; Raleigh, J.M.; Nag, N.; McLean, C.; Lelic, A.; Angel, C.; Guitera, P. High-risk cutaneous squamous cell carcinoma viral subtyping reveals differential expression of immune-related genes. Mod. Pathol. 2023, 36, 100010. [Google Scholar]
- Vygovska, M.; Hoyt, D.; Snyder, A.M.; Jonmundsson, T.; Khouri, A.; Sahni, D.R.; Ungar, J.; Lewin, J.M.; Gulati, N.; Phelps, R.G.; et al. Incidence and outcomes of Merkel cell carcinoma related to Merkel cell polyomavirus status in Iceland in 1981–2023. JAAD Int. 2024, 17, 192–199. [Google Scholar] [CrossRef]
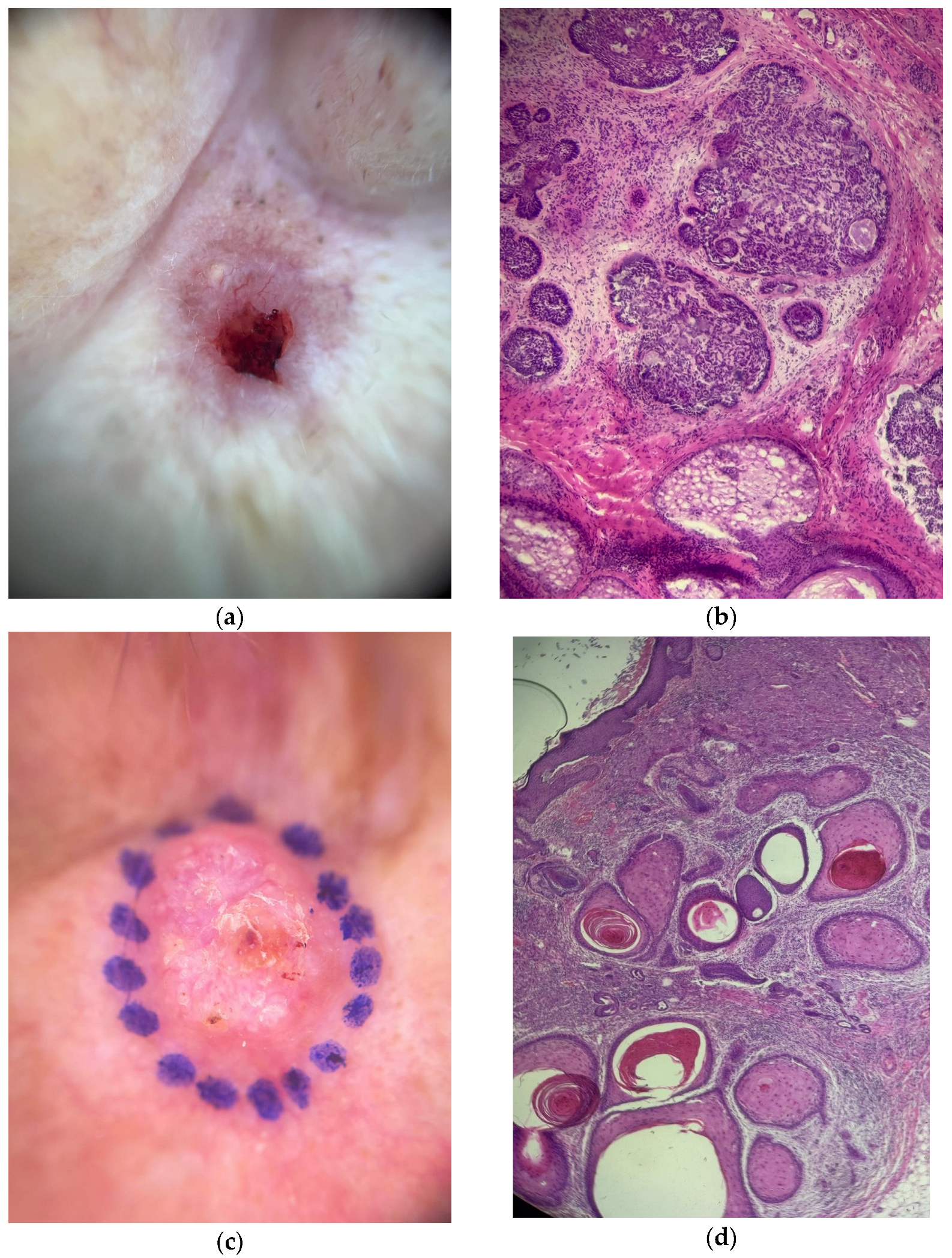
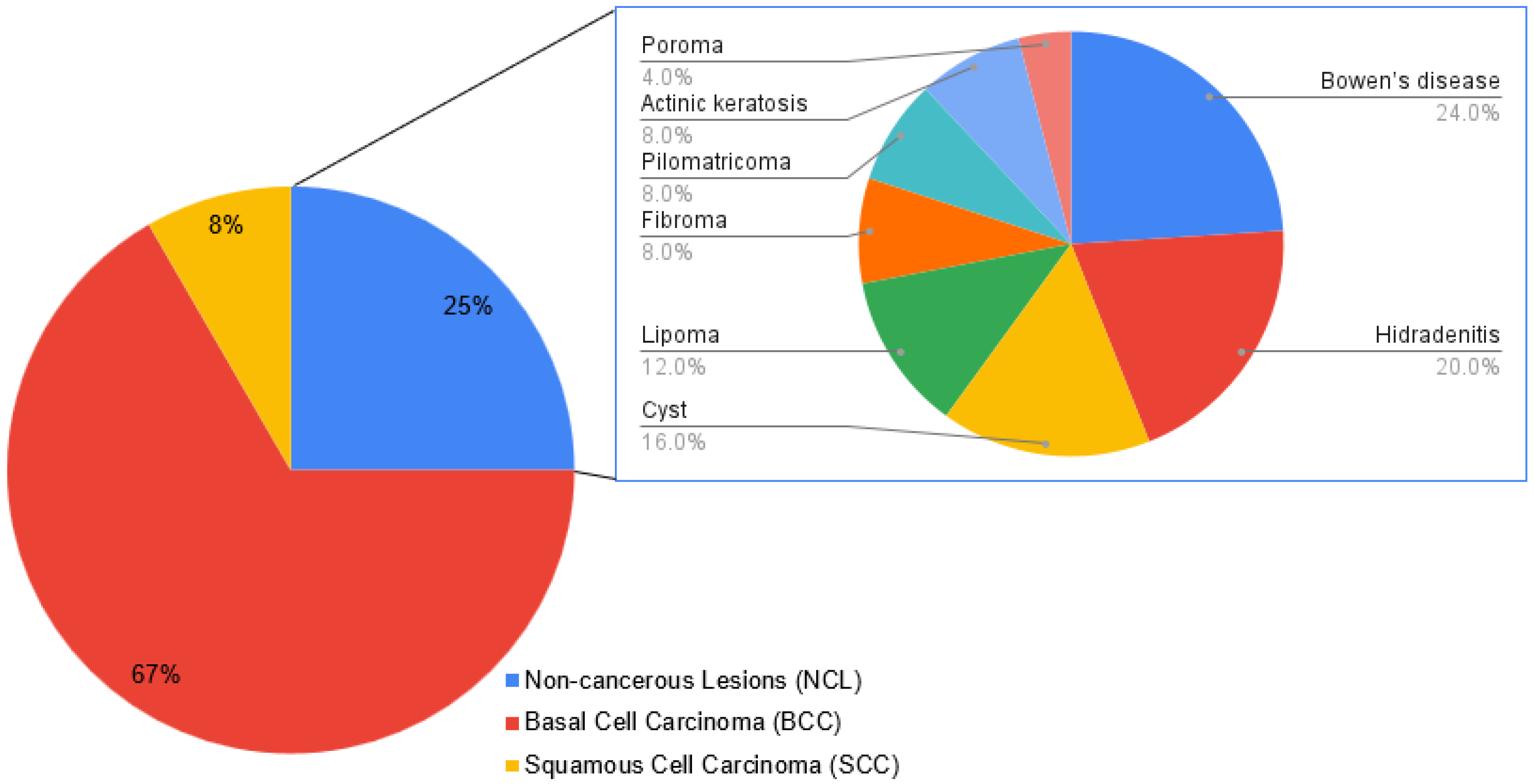


| Variable | n (%) | MCPyV Detection (%) | p Value |
|---|---|---|---|
| Gender | |||
| Male | 44 (52.4) | 53.2 | 0.224 |
| Female | 40 (47.6) | 46.8 | |
| Ethnicity | |||
| White | 70 (85.4) | 39.7 | 0.749 |
| Non-white | 14 (14.6) | 45.5 | |
| Age [mean yr. (range)] | 67 (17–91) | - | 0.354 |
| Skin lesion | |||
| BCC | 56 (66.7) | 38.2 | 0.674 |
| SCC | 7 (8.3) | 42.9 | |
| NCL | 21 (25) | 50 |
| Category | MCPyV+ | MCPyV− |
|---|---|---|
| Solar exposure | ||
| Low | 4 (25.0%) | 12 (75.0%) |
| Moderate | 8 (29.6%) | 19 (70.4%) |
| High | 45 (42.9%) | 60 (57.1%) |
| Type of sample | ||
| Surgical margin | 21 (23.3%) | 69 (76.7%) |
| Healthy skin | 16 (31.4%) | 35 (68.6%) |
| NMSC a | 33 (37.5%) | 55 (62.5%) |
| NCL b | 11 (44.0%) | 14 (56.0%) |
| Paired concordance c | ||
| Overall agreement | 13 (15.1%) | 47 (54.7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza, L.R.A.; Baez, C.F.; Bellott, T.R.; Pereira, M.S.; Gonçalves, M.T.V.; Guimarães, M.A.A.M.; Luz, F.B.; Varella, R.B. Increased Detection of Merkel Cell Polyomavirus in Non-Melanoma Skin Cancer and Its Association with Host Immunogenetic Profile. Dermato 2025, 5, 14. https://doi.org/10.3390/dermato5030014
de Souza LRA, Baez CF, Bellott TR, Pereira MS, Gonçalves MTV, Guimarães MAAM, Luz FB, Varella RB. Increased Detection of Merkel Cell Polyomavirus in Non-Melanoma Skin Cancer and Its Association with Host Immunogenetic Profile. Dermato. 2025; 5(3):14. https://doi.org/10.3390/dermato5030014
Chicago/Turabian Stylede Souza, Leonardo Ribeiro Alves, Camila Freze Baez, Thiago Rubim Bellott, Milena Siqueira Pereira, Marianna Tavares Venceslau Gonçalves, Maria Angelica Arpon Marandino Guimarães, Flávio Barbosa Luz, and Rafael Brandão Varella. 2025. "Increased Detection of Merkel Cell Polyomavirus in Non-Melanoma Skin Cancer and Its Association with Host Immunogenetic Profile" Dermato 5, no. 3: 14. https://doi.org/10.3390/dermato5030014
APA Stylede Souza, L. R. A., Baez, C. F., Bellott, T. R., Pereira, M. S., Gonçalves, M. T. V., Guimarães, M. A. A. M., Luz, F. B., & Varella, R. B. (2025). Increased Detection of Merkel Cell Polyomavirus in Non-Melanoma Skin Cancer and Its Association with Host Immunogenetic Profile. Dermato, 5(3), 14. https://doi.org/10.3390/dermato5030014







