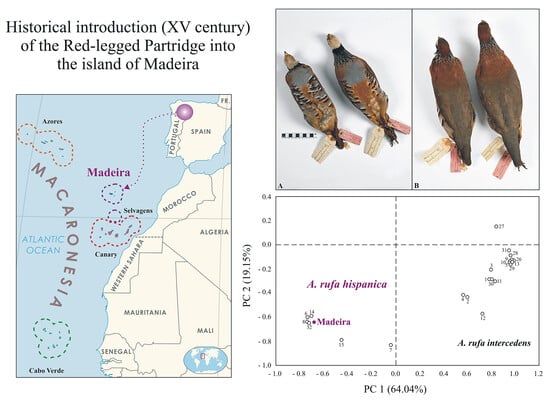
Genetic Identity of the Red-legged Partridge (Alectoris rufa, Phasianidae) from the Island of Madeira
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
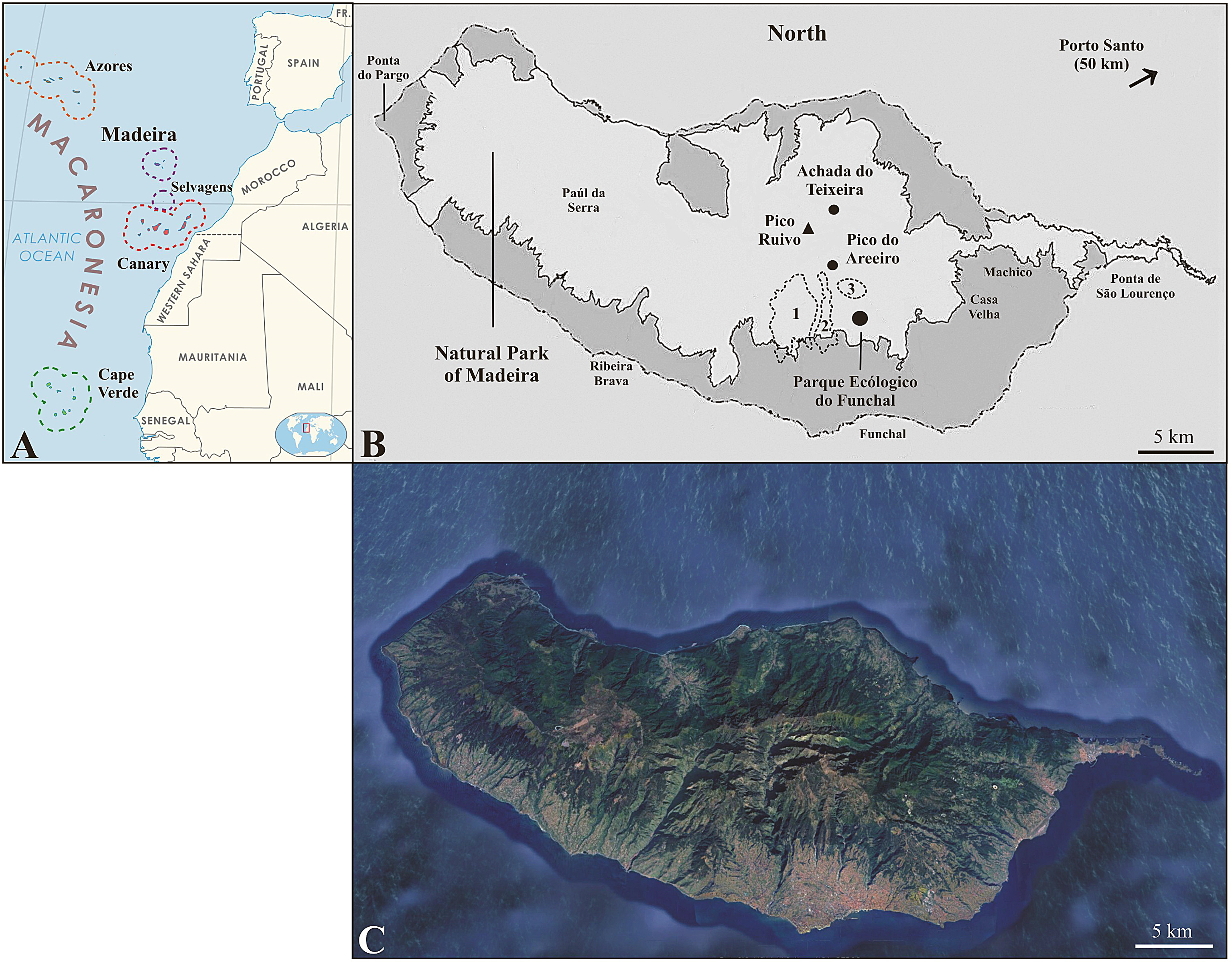
2.1. Study Area
2.2. Biological Sampling
2.3. DNA Extraction
2.4. Genetic Analysis and Data Elaboration
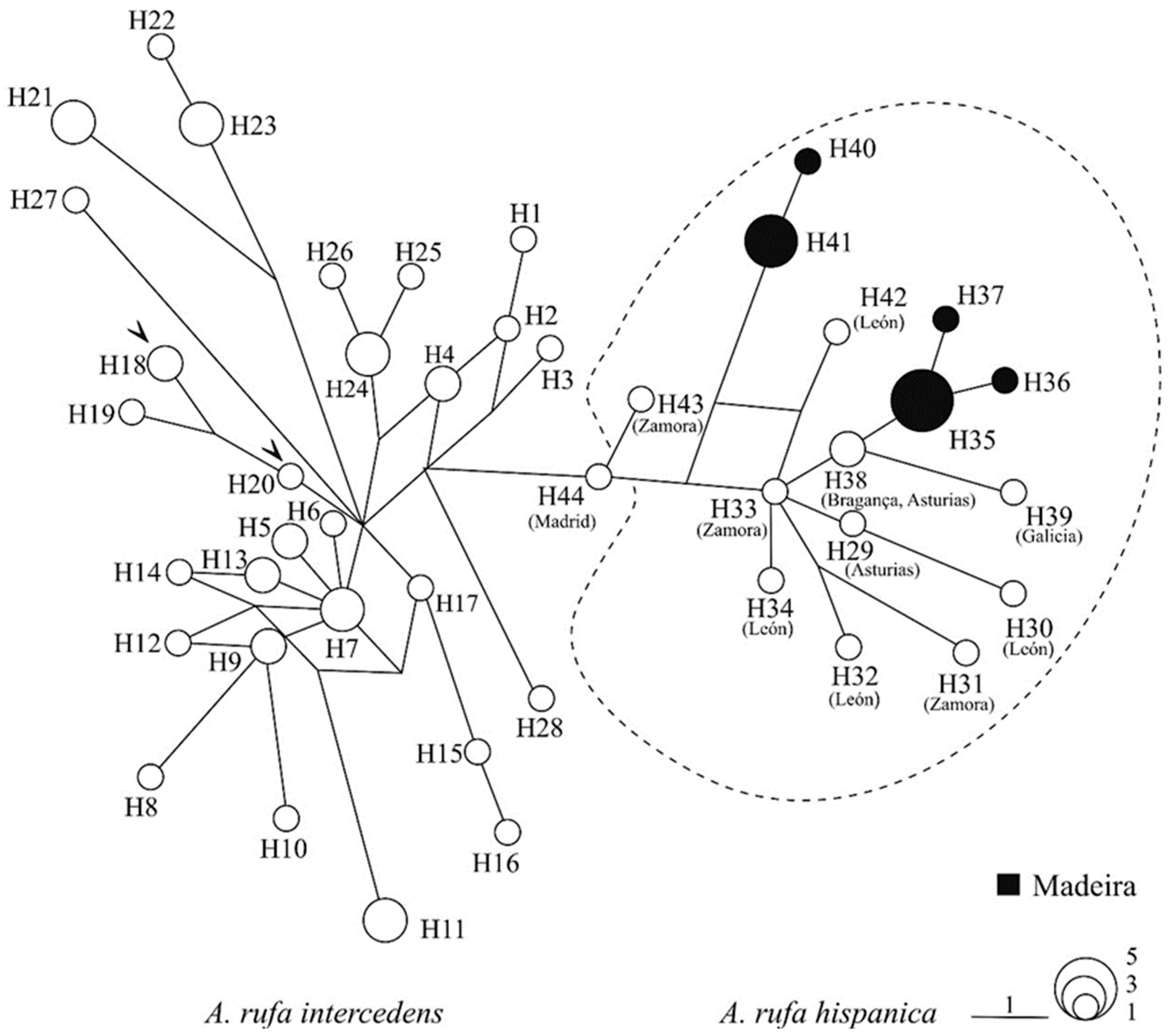
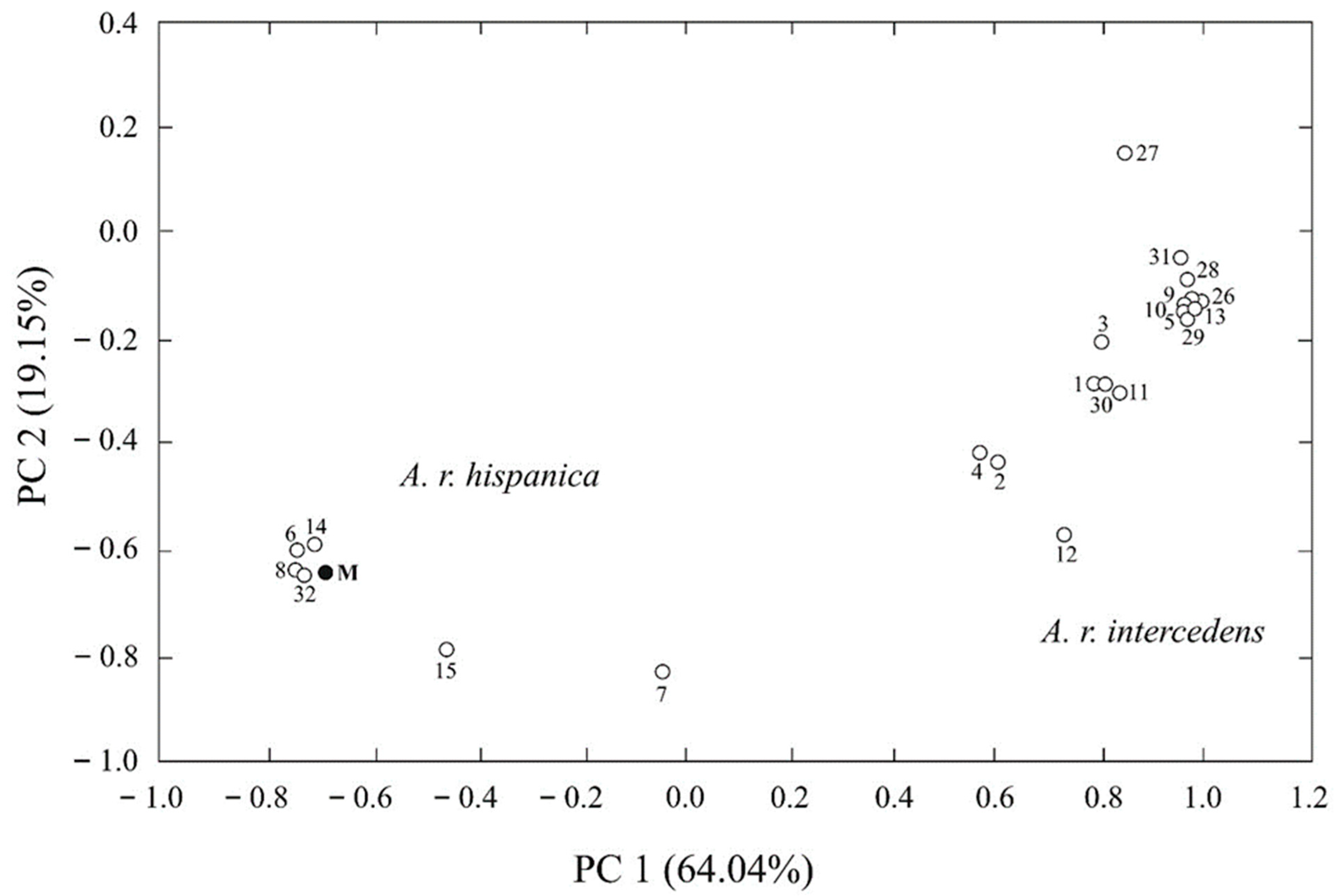
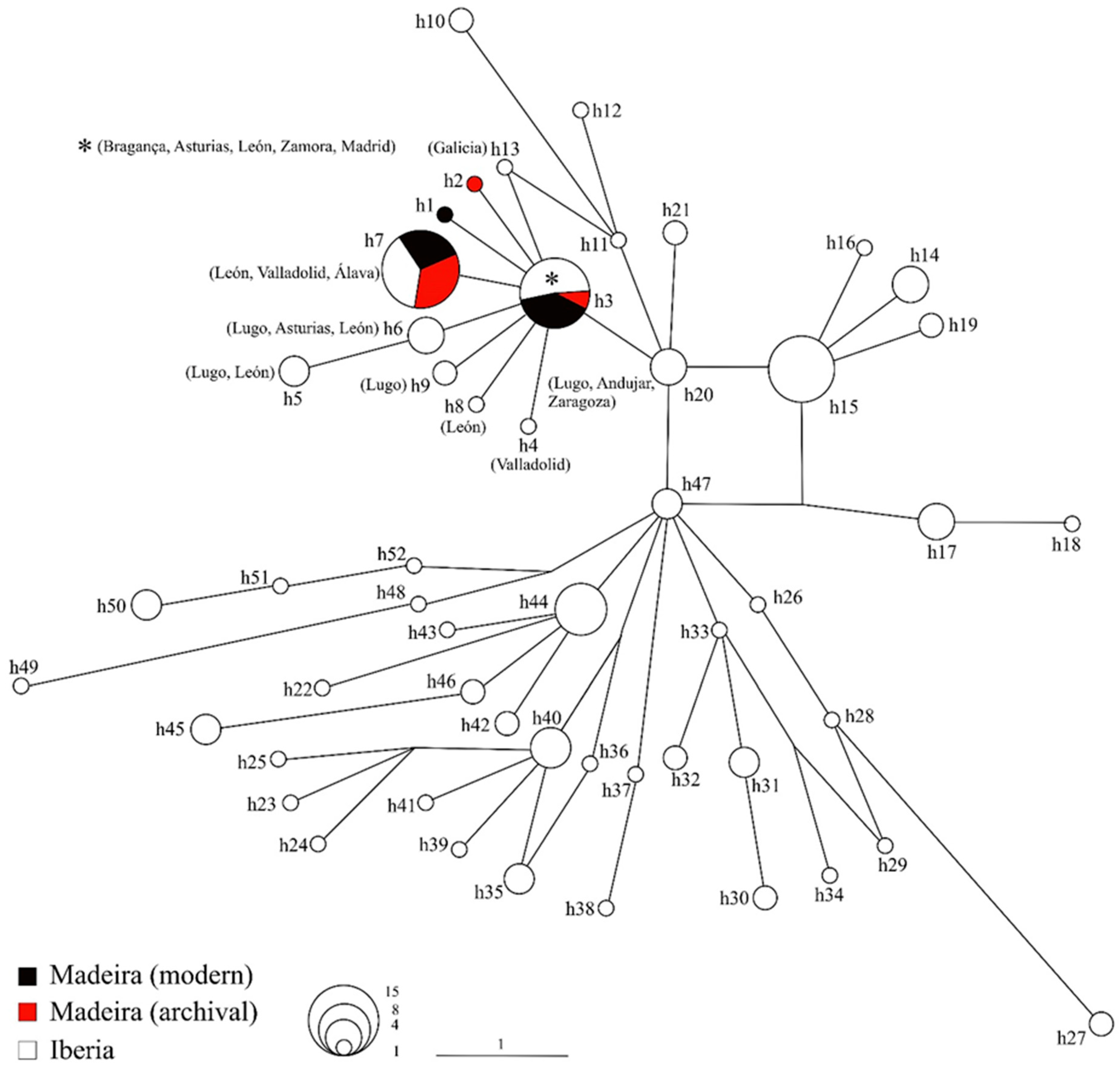
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clavero, M.; Sempere Marín, A. Effective cooperation between ecologists and historians for conservation: Documenting a 16th century crayfish introduction. Biol. Conserv. 2025, 11, 111405. [Google Scholar] [CrossRef]
- Johnsgard, P.A. The Quails, Partridges and Francolins of the World; Oxford University Press: Oxford, UK, 1988. [Google Scholar]
- Arnott, W.G. Birds in the Ancient World, From A to Z; Routledge: London, UK; New York, NJ, USA, 2007. [Google Scholar]
- Kabasakal, B.; Doğru, H.; Erdoğan, A.; Kaya, S. Transcontinental evolutionary dynamics and phylogeography of Alectoris (Aves: Galliformes): Identifying refugia, dispersal corridors, and cryptic diversity in the Palearctic region. Avian Res. 2025, 16, 100262. [Google Scholar] [CrossRef]
- Masseti, M. Representations of birds in Minoan art. Int. J. Osteoarchaeol. 1997, 7, 354–363. [Google Scholar] [CrossRef]
- Barbanera, F.; Marchi, C.; Guerrini, M.; Panayides, P.; Sokos, C.; Hadjigerou, P. Genetic structure of Mediterranean chukar (Alectoris chukar, Galliformes) populations: Conservation and management implications. Naturwissenschaften 2009, 96, 1203–1212. [Google Scholar] [CrossRef]
- Scandura, M.; Iacolina, L.; Apollonio, M.; Dessì-Fulgheri, F.; Baratti, M. Current status of the Sardinian partridge (Alectoris barbara) assessed by molecular markers. Eur. J. Wildl. Res. 2010, 56, 33–42. [Google Scholar] [CrossRef]
- Rodríguez Luengo, J.L. Fauna introducida. In Naturaleza de las Islas Canarias. Ecología y Conservación; Fernández Palacios, J.M., Martín Esquivel, J.L., Eds.; Editorial Turquesa: Santa Cruz, Spain, 2001; pp. 231–237. (In Spanish) [Google Scholar]
- McGowan, P.J.K.; Kirwan, G.M.; Boesman, P.F.D. Red-legged Partridge (Alectoris rufa), version 1.0. In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NT, USA, 2010. [Google Scholar]
- Villafuerte, R.; Negro, J.J. Digital imaging for colour measurement in ecological research. Ecol. Lett. 1998, 1, 151–154. [Google Scholar] [CrossRef]
- Barbanera, F.; Forcina, G.; Guerrini, G.; Dini, F. Molecular phylogeny and diversity of Corsican red-legged partridge: Hybridization and management issues. J. Zool. 2011, 285, 56–65. [Google Scholar] [CrossRef]
- Parrot, C. Caccabis rufa corsa subsp. nova. Ornithol. Monatsberichte 1910, 10, 156. [Google Scholar]
- Forcina, G.; Tang, Q.; Cros, E.; Guerrini, M.; Rheindt, F.E.; Barbanera, F. Genome-wide markers redeem the lost identity of a heavily managed gamebird. Proc. R. Soc. Lond. B Ser. 2021, 288, 20210285. [Google Scholar] [CrossRef]
- Barbanera, F. On the origins and history of the red-legged partridge (Alectoris rufa) from Elba Island (Tuscan Archipelago, Italy). Atti Soc. Tosc. Sci. Nat. Mem. Serie B 2021, 128, 45–55. [Google Scholar]
- Blanco-Aguiar, J.A.; Ferrero, E.; Dávila, J.A. Molecular DNA Studies in the Red-legged Partridge: From Population Genetics and Phylogeography to the Risk of Anthropogenic Hybridization. In The Future of the Red-Legged Partridge; Wildlife Research Monographs; Casas, F., García, J.T., Eds.; Springer: Cham, Germany, 2022; Volume 6, pp. 117–137. [Google Scholar]
- von Jordans, A. Alectoris rufa laubmanni . Novit. Zool. 1928, 34, 306. [Google Scholar]
- Cramp, S.; Simmons, K.E.L. Handbook of the Birds of Europe, the Middle East and North Africa. In The Birds of the Western Palearctic; Oxford University Press: Oxford, UK, 1980; Volume 2. [Google Scholar]
- Dickinson, E.C. The Howard and Moore Complete Check-List of the Birds of the World, 3rd ed.; Christopher Helm: London, UK, 2003. [Google Scholar]
- Barbanera, F.; Forcina, G.; Cappello, A.; Guerrini, M.; van Grouw, H.; Aebischer, N.J. Introductions over introductions: The genomic adulteration of an early genetically valuable alien species in the United Kingdom. Biol. Invasions 2015, 17, 409–422. [Google Scholar] [CrossRef]
- Barcelos, L.; Rodrigues, P.; Bried, J.; Mendonça, E.; Gabriel, R.; Borges, P. Birds from the Azores: An updated list with some comments on species distribution. Biodivers. Data J. 2015, 3, e6604. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T. Field Guide to the Birds of the Atlantic Islands; Christopher Helm: London, UK, 2006; pp. 1–368. [Google Scholar]
- Tristram, H.B. Caccabis rufa var. australis. In The Ibis; Series 6; British Ornithologists’ Union: Peterborough, UK, 1889; Volume 1, p. 28. [Google Scholar]
- Lever, C. Naturalised Birds of the World; T. & A. D. Poyser: London, UK, 2005; pp. 39–40. [Google Scholar]
- Tella, J.L. The unknown extent of ancient bird introductions. Ardeola 2011, 58, 399–404. [Google Scholar] [CrossRef]
- Bernström, J. Check-list of the breeding birds of the archipelago of Madeira. Bol. Mus. Mun. Funchal. 1951, 14, 64–82. [Google Scholar]
- Leite, J.D. Descobrimento da Ilha da Madeira e Discurso da Vida e Feitos dos Capitães da Dita Ilha: Tratado Composto em 1579 e Agora Publicado; Universidade de Coimbra: Coimbra, Portugal, 1947; pp. 1–157. (In Portuguese) [Google Scholar]
- da Montalboddo, F. Paesi Novamente Retrovati: Et Novo Mondo da Alberico Vespucio Florentino Intitulato; Enrico e Giovanni Maria Ca’ Zeno: Vicenza, Italy, 1507; pp. 1–119. (In Italian) [Google Scholar]
- Fructuoso, G. As Saudades da Terra. Historia das Hilas de Porto Sancto, Madeira, Desertas e Selvagens; Manuscripto do seculo XVI; Annotado por Alvaro Rodrigues de Azevedo, 1591. Typ; Funchalenses: Funchal, Portugal, 1873. (In Portuguese) [Google Scholar]
- Sloane, H. A Voyage to the Islands Madera, Barbados, Nieves, S. Christophers and Jamaica; Lehigh University: Bethlehem, PA, USA, 1707; Volume I, pp. 1–264. [Google Scholar]
- Bannerman, D.A.; Bannerman, W.M. Birds of the Atlantic Islands, A History of the Birds of Madeira, the Desertas, and the Porto Santo Islands; Oliver & Boyd: Edinburgh, UK, 1965; Volume II, pp. 1–207. [Google Scholar]
- Harcourt, E.V. A Sketch of Madeira; John Murray: London, UK, 1851; pp. 1–176. [Google Scholar]
- Zino, F.; Biscoito, M.; Zino, P.A. Birds of the archipelagos of Madeira and the Selvagens. New records and checklist. Bol. Mus. Munic. Funchal 1995, 47, 63–100. [Google Scholar]
- Tschusi zu Schmidhoffen, V. Caccabis rufa maderensis subsp. nov. Ornithol. Jahrb. 1904, 15, 106–108. [Google Scholar]
- Olden, J.D.; Rooney, T.P. On defining and quantifying biotic homogenization. Glob. Ecol. Biogeogr. 2006, 15, 113–120. [Google Scholar] [CrossRef]
- Fernández-Palacios, J.M.; Otto, R.; Capelo, J.; Caujapé-Castells, J.; de Nascimento, L.; Duarte, M.C.; Elias, R.B.; García-Verdugo, C.; Menezes de Sequeira, M.; Médail, F.; et al. In defence of the entity of Macaronesia as a biogeographical region. Biol. Rev. 2024, 99, 2060–2081. [Google Scholar] [CrossRef]
- Florencio, M.; Patiño, J.; Nogué, S.; Traveset, A.; Borges, P.A.V.; Schaefer, H.; Amorim, I.R.; Arnedo, M.; Ávila, S.P.; Cardoso, P.; et al. Macaronesia as a Fruitful Arena for Ecology, Evolution, and Conservation Biology. Front. Ecol. Evol. 2021, 9, 718169. [Google Scholar] [CrossRef]
- BirdLife International. European Red List of Birds; Publications Office of the European Union: Luxembourg, 2021; Available online: https://birdlifedata.blob.core.windows.net/sub-global/144_alectoris_rufa.pdf (accessed on 6 February 2025).
- Capelo, J.M.; Sequeira, M.; Jardim, R.; Costa, J.C. Guia da Excursão Geobotânica dos V Encontros ALFA 2004 à Ilha da Madeira. Quercetea 2004, 6, 5–45. (In Portuguese) [Google Scholar]
- Fontinha, S.; Henriques, D.; Nóbrega, H.; Teixeira, D.; Ferro, A.; Carvalho, M.A.A.P. Vegetation recovery after a large forest fire in the Ecological Park of Funchal (Madeira Island, Portugal). Silva Lusit. 2014, 22, 207–229. [Google Scholar]
- Barbanera, F.; Moretti, B.; Guerrini, M.; Al-Sheikhly, O.F.; Forcina, G. Investigation of ancient DNA to enhance natural history museum collections: Misidentification of smooth-coated otter (Lutrogale perspicillata) specimens across multiple museums. Belg. J. Zool. 2016, 146, 101–112. [Google Scholar] [CrossRef]
- Guerrini, M.; Barbanera, F. Non-invasive genotyping of the Red-legged Partridge (Alectoris rufa, Phasianidae): Semi-nested PCR of mitochondrial DNA from feces. Biochem. Genet. 2009, 47, 873–883. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Forcina, G.; Guerrini, M.; Barbanera, F. Non-native and hybrid in a changing environment: Conservation perspectives for the last Italian red-legged partridge (Alectoris rufa) population with long natural history. Zoology 2020, 138, 125740. [Google Scholar] [CrossRef]
- Ferrero, M.E.; Blanco-Aguiar, J.A.; Lougheed, S.C.; Sánchez-Barbudo, I.; de Nova, P.J.G.; Villafuerte, R.; Dávila, J.A. Phylogeography and genetic structure of the Red-legged Partridge (Alectoris rufa): More evidence for refugia within Iberian glacial refugium. Mol. Ecol. 2011, 20, 2628–2642. [Google Scholar] [CrossRef]
- Randi, E.; Lucchini, V. Organization and evolution of the mitochondrial DNA control region in the avian genus Alectoris. J. Mol. Evol. 1998, 47, 449–462. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of the nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Romano, H.; Correia-Fagundes, C.; Zino, F.; Biscoito, M. Birds of the archipelagos of Madeira and the Selvagens. II—New records and checklist update (1995–2010). Bol. Mus. Munic. Funchal 2010, 60, 5–44. [Google Scholar]
- Correia-Fagundes, C.; Romano, H.; Zino, F.; Biscoito, M. Birds of the archipelagos of Madeira and the Selvagens. III—New records and checklist update (2010–2020). Bol. Mus. Munic. Funchal 2021, 71, 5–20. [Google Scholar]
- Blanco-Aguiar, J.A.; Gonzalez-Jara, P.; Ferrero, M.E.; Sánchez-Barbudo, I.; Virgos, E.; Villafuerte, R.; Dávila, J.A. Assessment of game restocking contributions to anthropogenic hybridization: The case of the Iberian Red-legged Partridge. Anim. Conserv. 2008, 11, 535–545. [Google Scholar] [CrossRef]
- Barbanera, F.; Pergams, O.R.W.; Guerrini, M.; Forcina, G.; Panayides, P.; Dini, F. Genetic consequences of intensive management in game birds. Biol. Conserv. 2010, 143, 1259–1268. [Google Scholar] [CrossRef]
- Casas, F.; Mougeot, F.; Sánchez-Barbudo, I.; Dávila, J.A.; Viñuela, J. Fitness consequences of anthropogenic hybridization in wild Red-legged Partridge (Alectoris rufa, Phasianidae) populations. Biol. Invasions 2012, 14, 295–305. [Google Scholar] [CrossRef]
- Casas, F.; Mougeot, F.; Ferrero, M.E.; Sánchez-Barbudo, I.; Dávila, J.A.; Viñuela, J. Phenotypic differences in body size, body condition and circulating carotenoids between hybrid and “pure” Red-legged Partridges (Alectoris rufa) in the wild. J. Ornithol. 2013, 154, 803–811. [Google Scholar] [CrossRef]
- Dias, D. Rock (Alectoris graeca) and chukar (A. chukar) partridge introductions in Portugal and their possible hybridization with Red-legged Partridges (A. rufa): A research project. Gibier Faune Sauvag. 1992, 9, 781–784. [Google Scholar]
- Sánchez- García, C.; Sokos, C.; Santilli, F.; Ponce, F.; Sage, R.B.; Bro, E.; Buner, F.D. Enough Reared Red-Legs for Today, but Fewer Wild Ones for Tomorrow? The Dilemma of Gamebird Rearing and Releasing. In The Future of the Red-Legged Partridge; Wildlife Research Monographs, Casas, F., García, J.T., Eds.; Springer: Cham, Germany, 2022; Volume 6, pp. 139–173. [Google Scholar]
- Tanini, D.; Guerrini, M.; Vannini, C.; Barbanera, F. Unexpected genetic integrity boosts hope for the conservation of the Red-legged Partridge (Alectoris rufa, Galliformes) in Italy. Zoology 2022, 155, 126056. [Google Scholar] [CrossRef]
- Nunes de Sousa, P.J.F. A criação de perdiz-vermelha (Alectoris rufa) para repovoamento das serras da Madeira e Porto Santo. In 50 Anos a Servir a Floresta. Commemorações dos 50 Anos de Actividade Florestal, 1st ed.; Rocha da Silva, P.C., Soares de Sousa Carvalho, J.A., Nunes de Sousa, P.J.F., Freitas, P.J., Eds.; Governo Regional de Madeira, Secreteria Regional do Ambiente e Recursos Naturais: Funchal, Portugal; Direção Regional de Florestas: Funchal, Portugal, 2010; pp. 64–68. (In Portuguese) [Google Scholar]
- Caires, M. Direção de Florestas reduz produção de perdizes. In Diário de Notícias; Global Media Group: Lisbon, Portugal, 2012; p. 3. (In Portuguese) [Google Scholar]
- Funchal Noticias. Colónia de 124 Casais Reprodutores Garantem Anualmente Cerca de Dois Mil Exemplares de Perdiz Vermelha (In Portuguese). Available online: https://funchalnoticias.net/2016/03/08/secretaria-regional-do-ambiente-e-dos-recursos-naturais-reforca-especies-cinegeticas/ (accessed on 24 February 2025).
- Freitas, S.; Sousa, P.; Fernandes, J.P. Cinegética na ilha do Porto Santo: Usufruto social da biodiversidade insular. Bol. Mus. Munic. Funchal 2007, 12, 33–41. (In Portuguese) [Google Scholar]
- Peters, J.L. Check-List of Birds of the World; Harvard University Press: Cambridge, MA, USA, 1934; Volume II. [Google Scholar]
- Bump, G. Red-Legged Partridges of Spain; Special Scientific Report, Wildlife No. 39; United States Department of the Interior Fish and Wildlife Service: Washington, DC, USA, 1958; pp. 1–38.
- Carita, R. História da Madeira. Vol. I—Século XV—Matriz de Expansão Portuguesa; Imprensa Académica: Funchal, Portugal, 2014; pp. 1–239. (In Portuguese) [Google Scholar]
- Forcina, G.; Clavero, M.; Meister, M.; Barilaro, C.; Guerrini, M.; Barbanera, F. Introduced and extinct: Neglected archival specimens shed new light on the historical biogeography of an iconic avian species in the Mediterranean. Integr. Zool. 2024, 19, 887–897. [Google Scholar] [CrossRef]
- Baker, A.J.; Marshall, H.D. Mitochondrial Control Region Sequences as Tools for Understanding evolution. In Avian Molecular Evolution and Systematics; Mindell, D.P., Ed.; Academic Press: San Diego, CA, USA, 1997; pp. 51–82. [Google Scholar]
- Milberg, P.; Tyrberg, T. Naïve birds and noble savages—A review of man-caused prehistoric extinctions of island birds. Ecography 1993, 16, 229–250. [Google Scholar] [CrossRef]
- Forcina, G.; Panayides, P.; Kassinis, N.; Guerrini, M.; Barbanera, F. Genetic characterization of game bird island populations: The conservation of the black francolin (Francolinus francolinus) of Cyprus. J. Nat. Conserv. 2014, 22, 15–22. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerrini, M.; Berg, H.-M.; Frahnert, S.; Biscoito, M.; Barbanera, F. Genetic Identity of the Red-legged Partridge (Alectoris rufa, Phasianidae) from the Island of Madeira. Birds 2025, 6, 59. https://doi.org/10.3390/birds6040059
Guerrini M, Berg H-M, Frahnert S, Biscoito M, Barbanera F. Genetic Identity of the Red-legged Partridge (Alectoris rufa, Phasianidae) from the Island of Madeira. Birds. 2025; 6(4):59. https://doi.org/10.3390/birds6040059
Chicago/Turabian StyleGuerrini, Monica, Hans-Martin Berg, Sylke Frahnert, Manuel Biscoito, and Filippo Barbanera. 2025. "Genetic Identity of the Red-legged Partridge (Alectoris rufa, Phasianidae) from the Island of Madeira" Birds 6, no. 4: 59. https://doi.org/10.3390/birds6040059
APA StyleGuerrini, M., Berg, H.-M., Frahnert, S., Biscoito, M., & Barbanera, F. (2025). Genetic Identity of the Red-legged Partridge (Alectoris rufa, Phasianidae) from the Island of Madeira. Birds, 6(4), 59. https://doi.org/10.3390/birds6040059