Abstract
A severe consequence of SARS-CoV-2 infection that manifests as systemic inflammation and multi-organ involvement is called Multisystem Inflammatory Syndrome in Children (MIS-C). This review examines the possible relationship between gut barrier integrity, the microbiome, dysregulation of interleukin 6 (IL-6) signaling, and MIS-C. Clinical and biochemical features of MIS-C are comparable to those of other hyper-inflammatory syndromes, suggesting a dysregulated immune response. One possible explanation for the systemic inflammation seen in MIS-C patients is the SARS-CoV-2-induced dysregulation of the IL-6 signaling pathway. In addition, new data suggest a reciprocal link between gut barrier integrity and IL-6. SARS-CoV-2 exhibits bacteriophage-like behavior, highlighting the role of bacteria as a reservoir for the virus and emphasizing the importance of understanding the bacteriophagic mechanism of the virus in fecal–oral transmission. The increased translocation of viral products and bacterial toxins may result from disrupting the intestinal barrier and cause systemic inflammation. On the other hand, systemic inflammation can weaken the integrity of the intestinal barrier, which feeds back into the loop of immunological dysregulation. In the context of MIS-C, understanding the interaction between SARS-CoV-2 infection, IL-6, and gut barrier integrity may shed light on the etiology of the disease and guide treatment options. Since children with gut dysbiosis may be more susceptible to MIS-C, it is critical to reinforce their microbiome through probiotics supplementation, and plant-fiber-rich diets (prebiotics). Early antibiotic treatment and the use of zonulin antagonists should also be considered.
1. Introduction
COVID-19 is not as severe in children as it is in adults in most situations. Children are the target of around 18% of coronavirus infections overall and 1% of COVID-19 cases. The 5–15 age group has a higher incidence of COVID-19. Children in the United States have a seropositivity rate of approximately 75%, which is greater than that of adults. Fortunately, the majority of pediatric patients (>90%) with SARS-CoV-2 infection are asymptomatic or only show moderate signs such as weakness, dry cough, and low fever. Inpatient and outpatient records from 2021 revealed that only 2% of all infected children in the United States required intensive care unit (ICU) care. A total of 27% had moderate symptoms similar to influenza, 5% had substantial pneumonic symptoms, and 66% of all infected children exhibited no symptoms at all [1].
Reports from the UK appeared in May 2020 about children who needed to be admitted to intensive care units because of an unexplained multisystem inflammatory condition that resembled toxic shock syndrome and Kawasaki disease [2]. Subsequent reports of comparable cases were made in both Europe and the US, and they were linked to COVID-19 outbreaks both geographically and temporally [3,4,5]. While most afflicted youngsters tested positive for antibodies, indicating prior infection, RT-PCR results for the SARS-CoV-2 virus were negative. A SARS-CoV-2 infection-related post-infectious inflammatory response was suggested as the source of the clinical condition [6]. Similar cases pertaining to this novel syndrome were reported by the UK, the US, and the World Health Organization, and the illness temporarily associated with COVID-19 was referred to as multisystemic inflammatory syndrome in children (MIS-C) [6].
MIS-C is different from acute severe COVID-19 infection in children in terms of both epidemiological and clinical characteristics. Young age, a history of co-morbidity, respiratory symptoms, and respiratory dysfunction are linked to acute severe COVID-19 infection in children [7]. On the other hand, most of the MIS-C cases that were presented had considerable cardiovascular dysfunction and gastrointestinal (GI) symptoms. They were also older and did not often have co-morbidities. Such clinical characteristics match those identified in the most extensive and well-researched MIS-C case series available to date (n = 99) [8]. According to that study, 63% of children had cardiovascular impairment and 80% of children were between the ages of 6 and 12 years [8].
In a review work, Banoun [9] reiterated the importance of the gut microbiome (GM) in the severity of COVID-19, as the GM and respiratory infections are strongly linked and may influence the host’s response to pneumonia. The synthesis of short-chain fatty acids (SCFAs), the control of systemic inflammation, the establishment of oral immunological tolerance via regulatory T cells (Tregs), and the management of extra-intestinal T cell populations are some of the potential pathways [10]. Therefore, this paper hypothesizes that gut dysbiosis may also influence the pathogenesis of the MIS-C and clinical outcomes in affected children and presents evidence to support this proposal.
2. MIS-C Clinical Characteristics
COVID-19 and MIS-C are two distinct disorders caused by the SARS-CoV-2 virus [11]. MIS-C is an uncommon illness that may occur in children infected with SARS-CoV-2. It involves inflammation in various organs, including the brain, skin, eyes, heart, lungs, kidneys, and gastrointestinal tract. In total, 100% of the eight patients from the United Kingdom in the first correspondence to be published about MIS-C had GI issues [2]. Similarly, in an Italian study, GI problems affected 6 out of 10 children [4]. GI symptoms were reported in less than 10% to 15% of adult patients, but respiratory symptoms are the most prevalent presentation for them [12,13]. According to a study conducted in the USA, GI symptoms were reported in 84.1% of 44 children and were most frequently associated with rash (70.5%) and fever (100%) [14]. Other research found that 90% of 72 children had GI manifestations [15]. Although it can be life-threatening, most children recover with medical attention [11,16,17]. MIS-C was initially diagnosed as a form of Kawasaki disease due to their clinical similarities; however, subsequent research demonstrated that they are different entities with distinct epidemiological, clinical, and immunological profiles [18,19]. According to a recent study on GI involvement linked to COVID-19, excrement from up to 41% of children without MIS-C tested positive for SARS-CoV-2 [20]. This result suggested that the elevated inflammatory response is probably the cause of the high frequency of GI symptoms in MIS-C cases [15]. This work proposes an inverse relationship: SARS-CoV-2 infects and replicates within gut bacteria. Afterwards, these bacteria are destroyed and release toxic molecules that together with elevated IL-6 levels cause gut barrier dysfunction, leading to leakage of viral antigens and bacterial toxins to the blood. In other words, the hyper-inflammatory response observed in MIS-C is not the cause but rather the consequence of the immune system response against viral and bacterial toxins. In all the mechanisms so far proposed in the literature, although some authors highlight the persistence and presence of the virus in all the different forms including COVID-19, MIS-C, and long COVID, no model has considered that individual, familial, or community variability in the microbiome may generate different toxin-like peptides dependent on its bacterial genetic content. This element would explain the diversity of some symptoms in some individuals in the presence of the same viral infection.
3. The GM Supports Barrier Protection Functionality
The pathophysiology of numerous inflammatory and immunological disorders is intimately linked to the integrity of the intestinal barrier [21,22,23], a dynamic structure that interacts with and responds to a range of stimuli. It is composed of surface mucus, the epithelial layer, and immunological defenses [22]. The mucosal and epithelial components of the physical barrier are closely associated with several cellular junctions, such as adherens junctions (AJs), tight junctions (TJs), and desmosomes [24]. Additionally, the normal GM controls the intestinal micro-ecological equilibrium [25]. The intestinal barrier typically blocks substances and microbes from moving from the lumen to the circulation. However, intestinal dysbiosis, or the dysregulation of the gut flora, may result in a disorder called “leaky gut syndrome”, characterized by increased permeability that may trigger the innate immune system and promote low-grade inflammation. In recent times, GM dysbiosis has been linked to extra-intestinal as well as intestinal diseases. These include chronic diseases that are particularly prevalent in the elderly, such as diabetes [26,27,28], and other systemic side effects, such as oxidative stress, and increased inflammation [21,29,30].
4. The Link between Gut Microbiome Dysbiosis and MIS-C
Constant fever, vomiting, diarrhea, skin irritation, abdominal pain, and, in severe cases, hypotension and shock are the hallmarks of MIS-C [11,16,31]. One of the most common MIS-C symptoms is GI distress, raising the possibility that the GM may act as both a local and systemic inflammatory modulator [32,33]. The composition and activity of the GM have co-evolved with the host. It is influenced throughout life, from birth to old age, by a dynamic and complex interaction between the host genome and lifestyle variables, particularly nutrition, which is becoming recognized as the primary modulator of microbial activity [34,35].
This microbial population produces vitamins and ferments macromolecules including proteins, lipids, and carbohydrates. These metabolic effects are collectively known as co-metabolism [36]. Furthermore, the GM is primarily responsible for maintaining host homeostasis through the “training” of host immunity and structural/protective actions against commensal pathogens, specifically safeguarding the intestinal barrier [36]. Eubiosis is maintained by the GM when it is healthy. In pathological circumstances, the GM experiences an unbalanced state of dysbiosis wherein there is either a decrease in beneficial commensals, a proliferation of opportunistic pathogens, or both [37].
In adults, it is increasingly recognized that the gut acts as a reservoir for SARS-CoV-2 [38], and that dysbiosis and GI barrier disruption induce inflammatory activation in severe COVID-19 [39,40]. Several investigations have revealed that GM is also affected in adult patients with long COVID [41,42,43]. One study found that GM dysbiosis continued for up to 30 days after disease resolution, which may be related to persistent symptoms of post-acute sequalae of COVID-19 (PASC), also referred to as long COVID [44]. It is known that angiotensin-converting enzyme 2 (ACE2) regulates the synthesis of neutral amino acid transporters in the gut [45], which regulate the composition of the GM and, ultimately, the immune responses in the body [46,47]. In COVID-19 patients who already had age-related disorders, ACE2 imbalance has been associated with poor outcomes (including increased disease severity and mortality rate) through its impact on intestinal dysbiosis [48].
In a 6-month follow-up, Liu et al. [43] confirmed that long COVID patients had different GM species than controls and a consistently decreased diversity. Remarkably, the dysbiosis pattern was different among those who initially had COVID-19 but did not have long COVID. In the latter subgroup, tiredness, respiratory, and neuropsychiatric problems were strongly correlated with increased fecal relative abundance of opportunistic microorganisms [43]. Additional research revealed that long COVID is associated with dysbiosis of the GM in patients who have recovered a year after being discharged, suggesting that the GM may be crucial in long COVID [49].
Although the aforementioned findings explicitly demonstrated that alterations in the microbiome have a deleterious effect on the extended clinical course of COVID-19 in adults, there has been a lack of research on this crucial topic in MIS-C. The composition, diversity, and abundance of the gut microbiome were found to be different in MIS-C cases compared to COVID-19 cases and healthy controls. At the phylum level, the MIS-C group had an abundance of Bacteroidetes, whereas the healthy children had Firmicutes. When MIS-C was compared to the SARS-CoV-2 group and the healthy control group, the relative abundance of Bacteroidetes increased, and the Firmicutes/Bacteroides ratio significantly dropped [50]. Romani et al. [51] investigated the GM of children with COVID-19, including four cases of MIS-C. Veillonella, Ruminococcus, Clostridium, Dialister, and Streptococcus were more prevalent in the GM of MIS-C patients, while Bifidobacterium, Blautia, Granulicatella, and Prevotella were less prevalent.
Pro-inflammatory taxa were more prevalent and anti-inflammatory taxa were less prevalent in children with MIS-C. Children with MIS-C and COVID-19 [50] had lower concentrations of Faecalibacterium prausnitzii, which has been demonstrated to use butyrate to reduce intestinal mucosal inflammation and maintain gut physiology [52,53]. Therefore, it has been proposed that “alterations of the intestinal microbiome may contribute to the pathophysiology of MIS-C predisposing factors” [50].
4.1. SARS-CoV-2 Infection Impairs Gut Barrier Integrity by Inducing Zonulin Release
Intestinal bacteria are secluded from the underlying lamina propria by the dynamic physical and biochemical barrier known as the intestinal mucosa, which also inhibits the infiltration of pathogenic and antigenic molecules [54]. Despite the presence of chemical and physical barriers in the intestines, some pathogens can cross through it and interact with intestinal epithelial cells (IECs). Several sensing pattern recognition receptors (PRRs) are expressed by IECs, which are essential for maintaining the intestinal mucosal immune response’s homeostasis [55].
In response to bacterial signals, PRRs can initiate a physiological inflammatory response that promotes progenitor cell proliferation, epithelial cell survival, and pathogen elimination—all of which enhance intestinal integrity [56]. As was already mentioned, children with MIS-C frequently experience GI symptoms, which can lead to a severe hyper-inflammatory reaction and cardiac problems. A SARS-CoV-2 infection is known to cause MIS-C several weeks later, and it is known that the viral load in respiratory secretions decreases for 7–10 days following infection [57,58,59]. It seems doubtful that the initial respiratory tract infection in children with MIS-C is related to the disease, as the majority of them have negative nasopharyngeal viral swabs [19,57]. Furthermore, several lines of evidence demonstrated that SARS-CoV-2 can establish robust infection and replication in human IECs, which may contribute to GI dysfunction and potential fecal–oral transmission in some patients [60,61,62].
Zonulin is a member of a structurally and functionally similar family of proteins that modulate intercellular TJs to reversibly regulate intestinal permeability [63,64,65]. Large antigens, such as viral antigens generated from SARS-CoV-2 present in the GI tract, should not be able to pass through the gut lumen and into the bloodstream when the intestinal mucosal barrier is intact and functioning [38].
Patients with SARS-CoV-2 infection had elevated zonulin levels, which were linked to worse outcomes [33,66,67,68,69]. Furthermore, high serum zonulin levels disrupt the brain–blood barrier (BBB), allowing viruses to enter the brain and induce serious neurological symptoms [33,70]. Numerous autoimmune and hyper-inflammatory illnesses, including celiac disease [71], inflammatory bowel disease [72], and Kawasaki disease [73], have been linked to elevated circulating zonulin levels and increased intestinal permeability [72,74].
In 2021, Yonker et al. [33] examined bio-specimens from 100 children: 19 had acute COVID-19, 26 had MIS-C, and 55 were controls. Reverse transcription PCR (RT-PCR) was used to detect SARS-CoV-2 in stool samples, and zonulin and other indicators of mucosal barrier integrity were tested in plasma. According to this seminal research, the prolonged presence of undigested SARS-CoV-2 viruses causes the gastrointestinal tract to release more zonulin, which increases intestinal permeability [33].
Elevated zonulin levels suggest that intestinal epithelial TJs are failing, allowing SARS-CoV-2 antigens [33] and bacterial toxins [75,76] to penetrate the blood. It has been discovered that some strains of bacteria, mostly Gram-positive ones like Bifidobacterium and Lactobacillus spp., reduce intestinal zonulin levels while other strains, mostly Gram-negative ones like Salmonella spp., Prevotella, Escherichia coli, and Pseudomonas, induce zonulin release. These results are in line with earlier studies on animal models and cell lines (reviewed in [77]).
Several events, including gut dysbiosis [78], trigger the release of zonulin in a myeloid differentiation primary response 88 (MyD88)-dependent manner, which enables zonulin to bind to its target, protease-activated receptor 2 (PAR2), and subsequently transactivate the epidermal growth factor receptor (EGFR) [79]. This initiates a chain of events that results in the phosphorylation of TJ proteins, such as myosin 1c and zonula occludens 1 (ZO1), which in turn leads to the disintegration of TJs and an increase in the permeability of paracellular membranes to macromolecules [80]. This, in turn, accelerates the trafficking of viral and bacterial antigens into the bloodstream (Figure 1), resulting in hyper-inflammation [33].
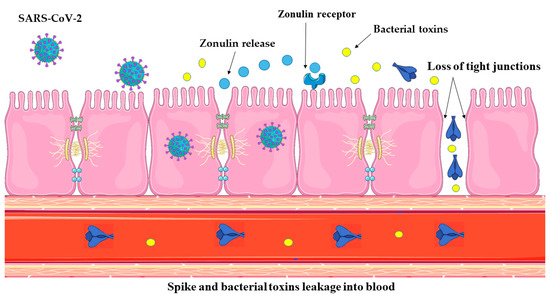
Figure 1.
Zonulin is a molecular gatekeeper that regulates TJs between intestinal epithelial cells. Normally, these TJs serve as a selective barrier, preventing chemicals and pathogens from entering the bloodstream from the intestines. SARS-CoV-2 infection in the GI tract causes localized inflammation of the mucosa, which in turn triggers zonulin release. This, in turn, enhances gut permeability, allowing SARS-CoV-2 antigens and bacterial toxins to pass through mucosal barriers and enter the bloodstream. Parts of the figure were drawn using pictures from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
The development of zonulin-dependent loss of gut integrity in infants affected with MIS-C suggests that chronic SARS-CoV-2 infection causes gut dysbiosis, leading to a progressive deterioration of mucosal barrier integrity. Viremia was not found in MIS-C [57], although antigenemia and viremia have been demonstrated to correlate with severe acute COVID-19 in adults [59,81].
Yonker et al. [33] found SARS-CoV-2 cleaved soluble components of spike (S1) and nucleocapsid antigens in the blood of children suffering from MIS-C, even though the children had been exposed to or had been infected with the virus weeks before. Compared to MIS-C patients and healthy controls (p < 0.0001), and children with acute COVID-19 (p < 0.001), SARS-CoV-2 spike protein levels were significantly higher. They also found that patients with MIS-C had significantly higher levels of SARS-CoV-2 S1 protein than healthy controls (p = 0.004) and children with acute COVID-19 (p = 0.02). Compared with healthy controls, the researchers found no discernible increase in blood levels of the SARS-CoV-2 antigen in children with acute COVID-19 [33].
It is known that a significant amount of blood that exits the gastrointestinal tract goes via the hepatic portal vein in the liver before entering the systemic circulation. As a result, it has long been understood that the liver is in control of immune surveillance against antigens coming from the gut [82]. It is important to mention that SARS-CoV-2 infection has been shown to impair liver immunity through several mechanisms, leading to liver injury and dysfunction. Hepatobiliary epithelial cells, cholangiocytes in particular, express the ACE2 receptor, which allows SARS-CoV-2 to infect these cells [83]. This direct infection can aggravate liver failure as the virus has been found in COVID-19 patients’ liver tissues, suggesting that this organ may be a target for viral replication [84,85]. A cytokine storm may result from the exacerbated immunological response that the infection sets off, which is defined by the release of pro-inflammatory cytokines. The liver’s capacity to develop a successful immune response may be hampered by this systemic inflammation, aggravating liver dysfunction and damage in adults [86] and in children [87]. According to a 2020 study, younger children—those under three years old—were more likely than older children to develop liver damage from COVID-19. The likely cause was early-life liver immaturity [88]. Upon admission, children frequently had higher liver enzyme levels than adults, who typically had an increase in enzyme levels no earlier than the second week of hospitalization [89]. Similar findings were reported in other investigations, with MIS-C showing these findings more frequently than in COVID-19 [87]. Therefore, the liver immune surveillance against antigens coming from the gut may be compromised in MIS-C, allowing the antigens to enter systemic circulation.
4.2. Bacteriophage-like Behavior of SARS-CoV-2 and MIS-C
Other important research performed by Brogna and colleagues revealed that this virus is capable of infecting intestinal bacteria, acting like a bacteriophage [90,91,92,93,94]. According to a previous study by that group, the blood, feces, and urine of COVID-19 patients contained toxin-like peptides that were almost identical to the poisonous elements of animal venoms, including conotoxin, phospholipases A2, phosphodiesterases, zinc metal proteinases, and bradykinins [75]. Later research revealed that SARS-CoV-2 might replicate autonomously in bacterial cultures derived from patient feces for up to 30 days and longer [95]. Viral-like structures ranging in size from 25 to 100 nm were observed by electron microscopy interacting with and within bacterial cell walls (Figure 2). The presence of SARS-CoV-2 nucleocapsid protein both inside and outside of the bacteria was verified by immunolabeling, specifically in two anti-inflammatory gut bacterial species normally present in a healthy human GM (Faecalibacterium prausnitzii and Dorea formicigenerans) [92]. These species were previously found to be considerably reduced in severe COVID-19 cases [96,97] and in children with MIS-C [50]. Importantly, both bacteria have been shown to use butyrate, thereby preserving gut physiology and reducing gut mucosal inflammation [52,53].
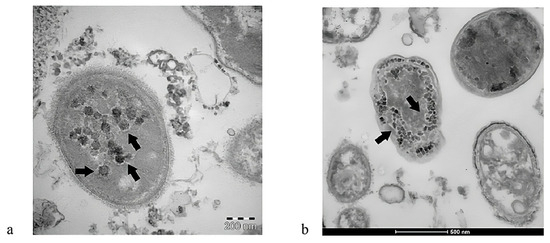
Figure 2.
SARS-CoV-2 can infect intestinal bacteria, thus showing a bacteriophage-like behavior. Panels (a,b): Images from a transmission electron microscope revealed the presence of SARS-CoV-2 inside two bacteria, marked by black arrows. Source: [91]. This figure is open access and is distributed under the Creative Commons Attribution Non-Commercial (CC BY-NC 4.0) license.
In addition, de novo synthesis of SARS-CoV-2 spike protein was detected in the bacterial cultures using 15N-labeled nitrogen as a source, accompanied by an increase in viral RNA load [92]. The discovery that SARS-CoV-2 can infect bacteria cells that are a component of the normal human GM and synthesize both nucleic acid and viral peptides has major consequences for human health, especially concerning the regulation and control of the virus spread. For example, the ability of SARS-CoV-2 to infect and replicate in human gut bacteria may lead to a scenario in which infected bacteria continue to harbor the virus for prolonged periods, possibly even after the systemic infection of the human host has resolved [92]. Epidemiological studies have shown that this may result in prolonged viral shedding through the intestinal tract, potentially increasing the likelihood of subsequent transmission. Furthermore, prolonged survival due to the continuous replication of the virus in the intestinal lumen could allow recurrent revival of systemic SARS-CoV-2 infection in the person, leading to COVID-19 relapse without the need for external reinfection [92].
The combined deleterious effects of SARS-CoV-2 infection reflected as increased zonulin release [33] and the decrease in commensal bacterial populations due to the bacteriophage-like effect of SARS-CoV-2 [90,91,92,93,94] could further compromise the integrity of the intestinal barrier, allowing the passage of the spike protein and toxins released by infected bacteria.
4.3. The Superantigen Hypothesis of SARS-CoV-2 Spike Protein
The proposed superantigen (Sag) activity of the SARS-CoV-2 spike protein has been a topic of investigation, particularly in relation to MIS-C. On one hand, the study by Sacco et al. [19] identified distinct immunopathologic signatures in pediatric COVID-19 and MIS-C and pointed out that MIS-C is characterized by prominent type II IFN and NF-κB-dependent signatures, matrisome activation, and increased levels of circulating spike protein. Only 2 of these 15 MIS-C patients showed a positive PCR on a nasopharyngeal swab within 7 days of the hospitalization, indicating that elevated spike protein levels were not caused by a prolonged respiratory tract infection, even though they did not look into the presence of SARS-CoV-2 mRNA in stool samples [19]. The study by Noval Rivas [98] suggests that “continuous and prolonged exposure to superantigen-like and neurotoxin-like viral motifs of the SARS-CoV-2 spike may promote autoimmunity leading to the development of post-acute COVID-19 syndromes, including MIS-C and long COVID, as well as neurological complications resulting from SARS-CoV-2 infection”. A study found that at the junction between the S1 and S2 subunits, the SARS-CoV-2 spike component S1 includes an insertion of four amino acids, P681RRA684 (PRRA), next to the cleavage site R685↑S686 [99]. Only SARS-CoV-2 and the β-coronaviruses in the SARS-like subfamily possess the polybasic segment PRRA [99]. The PRRA insert was discovered to be a component of the E661–R685 motif, a group of 25 amino acids with sequence and structural similarities to a portion of the SAg from SEB [100].
On the other hand, some studies also highlight that the SARS-CoV-2 spike protein does not exhibit superantigen-like activity [101,102]. In experimental setups, the spike protein was compared to staphylococcal enterotoxin B (SEB), a well-known superantigen. The study showed that while SEB induced a significant production of pro-inflammatory cytokines, the SARS-CoV-2 spike did not trigger a similar response in T cells, indicating that it lacks the intrinsic ability to function as a superantigen [101].
Another study examined the T cell response to SARS-CoV-2 peptide pools in children in the subacute phase of MIS-C in order to corroborate this observation. Despite their abundance, Vβ21.3-bearing CD4+ and CD8+ T cells did not belong to the SARS-CoV-2-specific T cell population. According to this research, most children’s T cell repertoire grew in response to an antigen unrelated to SARS-CoV-2 [103]. Although the antigen that triggers MIS-C is unknown, it might come from the associated leaky gut [33].
Burns [102] wrote: “If spike peptides are not superantigens, then what is the role of the spike protein in MIS-C pathogenesis?” Several groups reported the persistence of the spike antigen in the circulation of patients with acute MIS-C. However, Sigal et al. [104] used a sensitive electro-chemiluminescent immunoassay in patients with MIS-C and showed no persistence of the spike antigen in the plasma.
In 2022, reports from different countries reported a decrease in the prevalence of MIS-C in association with Omicron variant waves of SARS-CoV-2 infection. This questioned again the spike superantigen theory significance in the pathophysiology of MIS-C. Research conducted in 12 Israeli hospitals over a 16-week period during each of the three pandemic waves (Alpha, Delta, and Omicron) found that during the Omicron wave, there were fewer admissions to critical care and less severe cardiac outcomes. According to national data, there were 54.5 MIS-C incidents per 100,000 people under the age of 18 during Alpha, 49.2 during Delta, and 3.8 during Omicron. The reduced frequency of MIS-C during the Omicron wave may have been caused by vaccination, prior SARS-CoV-2 infection acting as a protective factor, or changes in the spike protein that resulted in diminished pathogenesis [105].
It is crucial to keep in mind that the furin-like cleavage site R685↑S686 is located next to this SAg-like motif [99]. Because the furin cleavage site (FCS) contains the SAg-like motif, it is essential for the acidic furin epitope to recognize it. In fact, viral mutations that eliminated the furin cleavage site have been shown to significantly decrease SARS-CoV-2-induced pathogenesis in the animal model [106]. It is important to clarify that the Delta variant did not lose the FCS. In that variant, a P681R mutation occurred near the FCS, and as a consequence, a glycosylation site was lost [107], leaving the FCS uncovered (Figure 3), thus promoting syncytia formation and a greater pathogenicity [108].
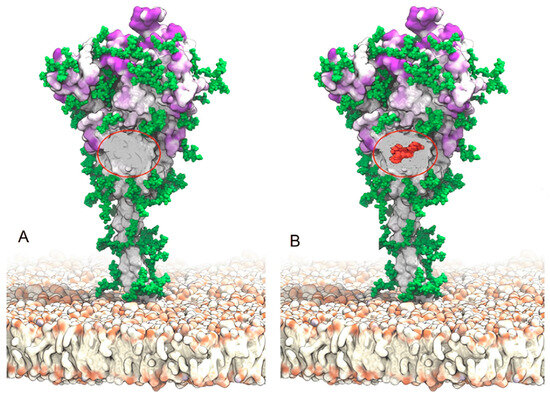
Figure 3.
(A) Spike protein from the Delta variant displaying the location of the FCS and the glycan hole (a region with no glycans indicated by a red oval). There were glycans covering this specific area in the original Wuhan strain. The Delta variant’s P681 mutation, on the other hand, caused glycans to be lost, which increased cell-to-cell fusion and syncytia formation, with increased pathogenicity as a result. (B) A Thr376 mutation in the Omicron variation led to the insertion of O-glycans (shown in red), whose structure blocks the FCS and prevents cell-to-cell fusion and the formation of syncytia. Source: [109]. This is an open-access article distributed under the Creative Commons Attribution Non-Commercial (CC BY-NC 4.0) license.
5. The Link between IL-6 Levels, Gut Barrier Integrity, and MIS-C
During the COVID-19 pandemic, several meta-analyses demonstrated that high levels of IL-6 have been linked to some adverse health effects, including acute respiratory distress syndrome, death, and ICU hospitalization. Serum IL-6 levels were almost three times greater in patients with such complex types of COVID-19 than in patients with less severe disease [110,111,112,113,114]. It has been demonstrated that the SARS-CoV-2 spike and nucleocapsid proteins alone can cause monocytes and macrophages to produce IL-6. Of interest, children with MIS-C exhibit hyper-phagocytosis and enhanced monocyte recruitment [19,115]. In certain COVID-19 patients, this IL-6 overexpression may be the catalyst that starts the dysregulated immune response [116,117]. However, extensive research allows us to understand that, depending on the specific circumstances of the immune response, IL-6 is a cytokine with multiple functions, having both pro- and anti-inflammatory effects [118]. Initially discovered as B cell stimulatory factor 2 (BSF-2), IL-6 stimulates activated B cells to produce immunoglobulin (Ig) [119]. At low, physiological concentrations, it regulates many important immune functions. Innate and adaptive immune systems are largely developed and activated in large part by IL-6. This cytokine induces monocytes to differentiate into macrophages rather than dendritic cells (DCs) in the innate immune system [120].
Moreover, the increase in anti-apoptotic proteins that support T cell survival is associated with IL-6 signaling [121,122,123]. Furthermore, IL-6 inhibits the transforming growth factor beta (TGF-β)-mediated maturation of Tregs and promotes the development of untrained CD4+ T cells into effector T cell subgroups, such as pathogen-specific effector Th17 cells [124,125]. Therefore, an increase in IL-6 levels causes immune dysregulation, as in COVID-19’s cytokine storm, and in other autoimmune disorders and cancer [124,126,127,128,129,130,131]. IL-6 is also essential for epithelial proliferation and wound repair [132] and for immune–epithelial–bacteria communication in both beneficial as well as harmful processes. For example, Pediococcus acidilactici K15, a lactic acid bacterial strain, promotes the secretion of IL-6 in BDCA1+ DCs (mDC1) via its double-stranded RNA and enhances the synthesis of protective IgA antibodies [133].
On the other hand, high levels of IL-6 can cause inflammation, and many probiotics work to lower these levels to treat intestinal disease [134]. In the context of ulcerative colitis (UC), a study found that at physiological levels, IL-6 controls epithelial barrier function by modulating the expression of TJ-related proteins. In contrast, IL-6 levels in the plasma of UC patients were elevated and increased as the disease worsened. The overproduction of IL-6 has also been shown to damage the intestinal epithelial cell barrier and control barrier function by increasing zonulin release. Conversely, when an anti-IL-6 antibody was added, the amount of zonulin was lower than in the control group [135].
It was discovered that long-term SARS-CoV-2 infection in the GI tract triggered zonulin release in children with MIS-C, with subsequent trafficking of SARS-CoV-2 antigens into the circulation, resulting in hyper-inflammation and signs of a cytokine storm, including significantly higher levels of tumor necrosis factor- α (TNF-α), IL-6, IL-10, and IL-1β [33]. Scientists demonstrated that SARS-CoV-2 RNA is still present in the GI tract for weeks after initial infection and viral antigenemia is correlated with zonulin-induced increased permeability (leaky gut) of the mucosal barrier [33].
Thus, it is likely that the high zonulin levels found in children with MIS-C [33] are due to excessive concentrations of IL-6 released by SARS-CoV-2-infected cells at the GI. In addition, several works have demonstrated that in MIS-C, impairment of the innate and adaptive immune responses is responsible for the prolonged presence of the virus in the GI tract [77,136,137,138,139,140].
6. Preventive and Therapeutic Strategies for MIS-C
Yonker et al. [33] used larazotide, a zonulin antagonist, to treat children with MIS-C and evaluated the impact on antigenemia and the children’s clinical outcomes. After receiving larazotide treatment for MIS-C, the patient’s plasma SARS-CoV-2 spike antigen levels and inflammatory markers decreased concurrently, leading to a greater clinical improvement than what was currently possible with known treatments. In adults with COVID-19 and long COVID, Brogna et al. [141] discovered that patients who started early antibiotic treatment showed a statistically significant decrease in recovery time, and such treatment was critical for maintaining high blood oxygen saturation levels. Delayed antibiotic initiation within the first 3 days increased the risk of pneumonia in both vaccinated and unvaccinated patients.
Furthermore, it is noteworthy that a considerable proportion of patients who were administered antibiotics within the initial three days and throughout the full seven days of the acute phase did not experience long COVID. One of the main contributing factors to the development of the disease appears to be the bacteriophage behavior of SARS-CoV-2 during the acute and post-COVID-19 phases. Early antibiotic treatment appears to be essential for halting the progression of disease, possibly controlling toxin release from infected bacteria, and preventing viral replication in the GM [141]. Severe streptococcal infections that mimic MIS-C were highlighted in a recent report [142]. Since MIS-C is an excluding diagnosis, the differential diagnosis needs to be carefully considered. Treatment is difficult for the physician because it overlaps with other prevalent disorders. Empirical antibiotic therapy for possible bacterial agents should be initiated in those with fever, organ involvement, and increased inflammatory markers. In fact, antibiotic therapy may be continued if clinical suspicion is high [142].
Another potential approach could involve employing anti-IL-6 antibodies such as tocilizumab or sarilumab. However, it has been shown that IL-6 suppression in COVID-19 patients affects the neutralizing capacity of anti-SARS-CoV-2 antibodies [143]. Considering that the neutralizing activity of anti-SARS-CoV-2 antibodies determines protection against symptomatic infection [144], the study conducted by Della-Torre calls for a rigorous re-evaluation of the risk of reinfection and severe illness in patients receiving anti-IL-6 antibodies [143]. It is also important to discuss the phenomena of COVID-19 rebound, which has been documented in individuals treated with antiviral medications after their initial infection [145]. In particular, it has been demonstrated that nirmatrelvir therapy during SARS-CoV-2 infection diminishes the likelihood of developing severe COVID-19, but it also inhibits the production of T cells and antibodies specific to SARS-CoV-2 [146]. This causes some patients’ viral loads to rebound and their COVID-19 symptoms to recur quickly following the completion of prompt and efficient nirmatrelvir treatment. Furthermore, the development of efficient long-term immunity may be hampered by this process [146].
Certain Gram-negative bacterial strains, such as Escherichia coli, Prevotella, Pseudomonas, and Salmonella spp., have been found to induce intestinal zonulin release, while other strains, primarily Gram-positive ones, like Bifidobacterium and Lactobacillus spp., have been found to decrease zonulin levels. These findings are consistent with previous research conducted on cell lines and animal models (reviewed in [147]). Probiotics are live microorganisms that, when administered in the right quantities and for the right length of time, benefit the health of the host [148]. Through their surface molecules and metabolites, probiotics and intestinal symbionts can alter the host’s intestinal barrier function [149]. Several studies have shown that probiotics reduce both intestinal permeability and epithelial barrier dysfunction in gastrointestinal disorders, thereby demonstrating the role of GM in improving intestinal barrier function and protection (Figure 4) against pathogens [150,151,152].
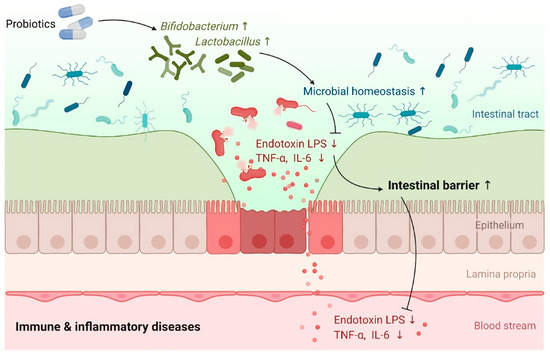
Figure 4.
A graphical representation shows the influence of probiotics on intestinal barrier function in immunological and inflammatory conditions. Source: [152]. This figure is open access and is distributed under the Creative Commons Attribution Non-Commercial (CC BY-NC 4.0) license.
At present, there is no doubt about the link between GM and the regulation of zonulin release, as some probiotic strains have been shown to improve gut barrier function by affecting the expression of zonulin and TJ proteins [147,153,154,155]. Probiotics dramatically enhanced gut barrier functioning, according to a meta-analysis of data from a total of 26 randomized controlled trials (n = 1891). Specifically, the trans-epithelial resistance (TER) was significantly enhanced, while serum zonulin, endotoxin, and lipopolysaccharide levels were importantly reduced. Moreover, probiotic groups outperformed control groups in lowering inflammatory markers like IL-6,TNF-α, and C reactive protein. Additionally, probiotics can regulate the composition of the GM by increasing the enrichment of Lactobacillus and Bifidobacterium [152].
Oral probiotic therapy has also been utilized in pediatric and adult populations to reduce the incidence and severity of respiratory infections. By strengthening the gut–lung axis and controlling the host inflammatory response, probiotics can elicit antiviral effects [156]. In a pediatric experiment (n = 31 prebiotics, 31 probiotics, and 32 placebos), preterm infants were given a prebiotic combination of galacto-oligosaccharide and polydextrose or the probiotic Lactobacillus rhamnosus GG mixed with breast milk throughout the first 60 days of life. There were fewer cases of virus-associated respiratory tract infections in the probiotic and prebiotic groups (p = 0.022 and p < 0.001, respectively) [157]. Probiotics may lessen symptoms of upper respiratory tract infections and stabilize GM diversity, according to a study that supplied Lab4P probiotics (comprising lactobacilli and bifidobacteria) daily to 220 overweight and obese adults [156,158]. This may be especially important for COVID-19 disease, where obesity is linked to worse outcomes [159,160].
Research into the treatment of COVID-19 in children using oral microbial interventions is scarce, despite its potential [32,161]. Probiotics have only been tested in a small number of adult patients with COVID-19, but the results were encouraging, showing reductions in viral load, hospitalization duration, death, and diarrhea frequency [161,162,163,164,165,166,167]. Adults with moderate-to-severe COVID-19 were given a supplementary oral dose of the Bifidobacterium animalis sp. Lactis strain (n = 20 probiotic, 24 non-probiotic). The probiotic group experienced a five-day reduction in hospital stay (p < 0.001) and a corresponding decrease in IL-6 levels (p < 0.001) [164].
A single-center, quadruple-blinded, randomized trial was carried out by other researchers on adult outpatients with symptomatic COVID-19. For 30 days, subjects were randomly assigned to either a probiotic formula (including Lactiplantibacillus plantarum KABP022, KABP023, and KAPB033, as well as Pediococcus acidilactici KABP021) or a placebo [163]. In total, 78 of 147 (53.1%) patients in the probiotic group accomplished complete remission, as opposed to 41 of 146 (28.1%) in the placebo group. There were no hospitalizations or deaths during the research, and the nasopharyngeal viral load, lung infiltrates, and duration of both digestive and non-digestive symptoms were all reduced when compared to the placebo. There were no significant differences in fecal microbiome composition between probiotic and placebo groups, although probiotic treatment boosted specific IgM and IgG antibodies against SARS-CoV-2 when compared to the placebo. Therefore, rather than altering the diversity of the colonic microbiome, it is assumed that probiotics predominantly enhance the host´s immune system [163]. They promote the synthesis of immunoglobulins, specifically IgA and type I interferons, and boost the induction of interleukins and the activation of macrophages, natural killer cells, and T-helper cells [168,169].
Probiotic supplementation may therefore help to restore gut health by enhancing immune responses, reducing inflammation, and reinforcing the epithelial barrier, thus possibly decreasing susceptibility to severe SARS-CoV-2 infection. It is suggested that probiotics should be used as a prophylactic rather than a treatment for severe cases of MIS-C, as once a cytokine storm is activated, it is very difficult to control.
7. Conclusions
A severe hyper-inflammatory illness known as MIS-C is linked to infection with the SARS-CoV-2 virus. After contracting COVID-19, children and adolescents can develop this uncommon but potentially dangerous syndrome, which is characterized by fever, increased inflammatory markers, and predominant gastrointestinal symptoms [6]. After the acute sickness passes, immunological dysregulation is thought to be the cause, with a genetic susceptibility in some cases [19]. Although the complex relationship between invasive viruses and host physiology is not yet fully understood, increasing data suggest that the GM may influence the course of viral diseases [169]. Research also identified autoantibodies against endothelial cells and cardiomyocytes, suggesting that MIS-C may be an autoimmune response triggered by the viral infection [170,171]. The superantigen hypothesis proposed that a domain within the SARS-CoV-2 protein could excessively stimulate the immune system and contribute to the hyper-inflammatory state observed in MIS-C [100]. However, experimental work demonstrated that the spike protein lacks superantigen activity [101].
This work has delved into the multifaceted interactions between MIS-C, elevated levels of IL-6 induced by SARS-CoV-2 infection, the microbiome, and gut barrier integrity. A significant aspect of our pathogenic model is the mechanism by which the virus impairs gut barrier integrity. In addition to the infection and destruction of beneficial bacteria by SARS-CoV-2 [90,91,92,93,94,95], this virus also induces an excessive IL-6-mediated zonulin release that increases intestinal permeability (leaky gut), so the spike protein and toxins released by the bacteria pass into the bloodstream, causing MIS-C [33]. Although some researchers discovered the presence of the spike antigen in blood from children with acute MIS-C, subsequent work showed no persistence of the spike antigen in the plasma, and more detailed analysis is needed to determine if the circulating spike protein in patients with MIS-C is involved in disease pathogenesis [104]. In our model, the initial pathogenic stimulus is provided by the virus inducing IL-6 release and damaging the gut barrier. SARS-CoV-2 infects and destroys beneficial bacteria in the gut, favoring the growth of pathogenic bacteria that release toxic products into the blood [90,92,93]. This probably triggers a hyper-inflammatory response resembling the toxic shock syndrome. Interestingly, the prevalence of beneficial bacteria like Bifidobacterium, Faecalibacterium prausnitzii, and Dorea formicigenerans was found to be much lower in severe COVID-19 cases [96,97], in PACS [43,44], and in children with MIS-C [50]. Such a reduction in the number of these bacteria could be due to the destruction caused by the lytic phase of SARS-CoV-2 [90].
The presence of the FCS located in the spike protein [99] could cause further damage to the integrity of the gut barrier. Research demonstrated that syncytium formation was related to severe disease outcomes in COVID-19, as it led to extensive tissue damage in the lungs. The ability of the virus to induce syncytia is also thought to contribute to the inflammatory response observed in infected patients, as the released S1 subunit from the spike protein can activate immune receptors, further exacerbating the disease [172,173]. Interestingly, in 2022, several countries reported a significant reduction in the incidence of MIS-C cases in association with Omicron waves. A study found that during the Omicron wave, there were fewer admissions to the critical care ward, with 54.5 MIS-C cases per 100,000 children under the age of 18 during Alpha, 49.2 during Delta, and 3.8 during Omicron [105]. We suggest that such reduced incidence was caused by a mutation that added O-glycans to the FCS in Omicron, since such mutation has been shown to significantly decrease SARS-CoV-2-induced pathogenesis in the animal model [106]. It is possible that the Alpha and Delta strains induced greater damage to the gut barrier by forming syncytia, thus causing an enhanced leakage of bacterial toxins to the blood.
Due to the acknowledged relevance of the GM in MIS-C, COVID-19, and long COVID, preventive and therapeutic strategies should prioritize restoring and maintaining GM balance and enhancing gut barrier integrity. There is evidence that if damage to the mucosal epithelium is prevented and treated early in the disease, MIS-C may not develop [33]. Given that gut dysbiosis may predispose children to MIS-C, it is important to strengthen their microbiome by consuming probiotics, and diets high in plant fiber (prebiotics). Therapeutic agents like larazotide [33] and antibiotics [95,141] constitute the best options. An experimental strategy was created to culture in vitro fecal microbiota from adult infected persons, monitor the presence of SARS-CoV-2, and compare the effects of various antibiotics. It was discovered that viral replication parallels bacterial growth and is affected by the use of particular antibiotics. In the four aliquots treated with metronidazole, vancomycin, amoxicillin, and azithromycin, respectively, the viral load was decreased to undetectable levels (100% efficacy), while Cefixime reduced viral load by 85%, ciprofloxacin by 61%, and teicoplanin by 56% [95]. Since metronidazole and amoxicillin are frequently used in children, they could be safely used to treat severe MIS-C cases.
The use of anti-IL-6 antibodies can lead to immune suppression, increasing the risk of infections [143]. The use of antivirals like nirmatrelvir could cause a COVID-19 rebound by inhibiting the production of T cells and antibodies specific to SARS-CoV-2, and hampering the development of efficient long-term immunity [145,146]. In conclusion, given that gut dysbiosis appears to be the predisposing factor to MIS-C development, strengthening the microbiome by consuming probiotics, particularly butyrate-producing Bifidobacterium, and diets high in plant fiber (prebiotics) should be a strategy for further research as a preventative measure, and the use of specific antibiotics to help restore the beneficial GM makeup post disease development to curb the viral ability to dysregulate the gut flora in afflicted children.
Author Contributions
Conceptualization, V.N.U., E.M.R. and A.R.-C.; formal analysis, A.Z., E.M.R., M.R., D.C., A.H.-J., V.N.U., M.F., C.B., M.P., and A.R.-C.; investigation, A.Z., E.M.R., M.R., D.C., A.H.-J., V.N.U., M.F., C.B., M.P., and A.R.-C.; data curation, A.Z., E.M.R., M.R., D.C., A.H.-J., V.N.U., M.F., C.B., M.P., and A.R.-C.; writing—original draft preparation, A.Z., E.M.R., M.R., D.C., A.H.-J., V.N.U., M.F., C.B., M.P., and A.R.-C.; writing—review and editing, A.Z., E.M.R., M.R., D.C., A.H.-J., V.N.U., M.F., C.B., M.P., and A.R.-C.; visualization, V.N.U., A.Z. and A.R.-C.; supervision, V.N.U., E.M.R. and A.R.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
Dr. Brogna Carlo is the scientific and technical director of the Craniomed group. No conflict of interest or economic participation is present in this or any other work. Dr. Mikolaj Raszec is the founder and managing editor of Merogenomics, the company performs genomic sequencing for people with undiagnosed diseases, cancer profiling and mothers for prenatal screening. Therefore, this author also declares no conflict of interest.
References
- Forrest, C.B.; Burrows, E.K.; Mejias, A.; Razzaghi, H.; Christakis, D.; Jhaveri, R.; Lee, G.M.; Pajor, N.M.; Rao, S.; Thacker, D. Severity of acute COVID-19 in children< 18 years old March 2020 to December 2021. Pediatrics 2022, 149, e2021055765. [Google Scholar] [PubMed]
- Riphagen, S.; Gomez, X.; Gonzalez-Martinez, C.; Wilkinson, N.; Theocharis, P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet 2020, 395, 1607–1608. [Google Scholar] [CrossRef] [PubMed]
- Dolinger, M.T.; Person, H.; Smith, R.; Jarchin, L.; Pittman, N.; Dubinsky, M.C.; Lai, J. Pediatric Crohn disease and multisystem inflammatory syndrome in children (MIS-C) and COVID-19 treated with infliximab. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 153–155. [Google Scholar] [CrossRef]
- Verdoni, L.; Mazza, A.; Gervasoni, A.; Martelli, L.; Ruggeri, M.; Ciuffreda, M.; Bonanomi, E.; D’Antiga, L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: An observational cohort study. Lancet 2020, 395, 1771–1778. [Google Scholar] [CrossRef]
- Chiotos, K.; Bassiri, H.; Behrens, E.M.; Blatz, A.M.; Chang, J.; Diorio, C.; Fitzgerald, J.C.; Topjian, A.; John, A.R.O. Multisystem inflammatory syndrome in children during the coronavirus 2019 pandemic: A case series. J. Pediatr. Infect. Dis. Soc. 2020, 9, 393–398. [Google Scholar] [CrossRef]
- Radia, T.; Williams, N.; Agrawal, P.; Harman, K.; Weale, J.; Cook, J.; Gupta, A. Multi-system inflammatory syndrome in children & adolescents (MIS-C): A systematic review of clinical features and presentation. Paediatr. Respir. Rev. 2021, 38, 51–57. [Google Scholar]
- Derespina, K.R.; Kaushik, S.; Plichta, A.; Conway Jr, E.E.; Bercow, A.; Choi, J.; Eisenberg, R.; Gillen, J.; Sen, A.I.; Hennigan, C.M. Clinical manifestations and outcomes of critically ill children and adolescents with coronavirus disease 2019 in New York City. J. Pediatr. 2020, 226, 55–63.e52. [Google Scholar] [CrossRef] [PubMed]
- Dufort, E.M.; Koumans, E.H.; Chow, E.J.; Rosenthal, E.M.; Muse, A.; Rowlands, J.; Barranco, M.A.; Maxted, A.M.; Rosenberg, E.S.; Easton, D. Multisystem inflammatory syndrome in children in New York State. N. Engl. J. Med. 2020, 383, 347–358. [Google Scholar] [CrossRef]
- Banoun, H. Why are children and many adults not affected by COVID-19? Role of the host immune response. Infect. Dis. Res. 2022, 3, 18. [Google Scholar] [CrossRef]
- Chunxi, L.; Haiyue, L.; Yanxia, L.; Jianbing, P.; Jin, S. The gut microbiota and respiratory diseases: New evidence. J. Immunol. Res. 2020, 2020. [Google Scholar] [CrossRef]
- Fraser, R.; Orta-Resendiz, A.; Dockrell, D.; Müller-Trutwin, M.; Mazein, A. Severe COVID-19 versus multisystem inflammatory syndrome: Comparing two critical outcomes of SARS-CoV-2 infection. Eur. Respir. Rev. 2023, 32. [Google Scholar] [CrossRef]
- Mao, R.; Qiu, Y.; He, J.-S.; Tan, J.-Y.; Li, X.-H.; Liang, J.; Shen, J.; Zhu, L.-R.; Chen, Y.; Iacucci, M. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.; Altayar, O.; Siddique, S.M.; Davitkov, P.; Feuerstein, J.D.; Lim, J.K.; Falck-Ytter, Y.; El-Serag, H.B.; Institute, A. AGA institute rapid review of the gastrointestinal and liver manifestations of COVID-19, meta-analysis of international data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology 2020, 159, 320–334.e327. [Google Scholar] [CrossRef]
- Miller, J.; Cantor, A.; Zachariah, P.; Ahn, D.; Martinez, M.; Margolis, K.G. Gastrointestinal symptoms as a major presentation component of a novel multisystem inflammatory syndrome in children that is related to coronavirus disease 2019: A single center experience of 44 cases. Gastroenterology 2020, 159, 1571–1574.e1572. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-H.; Kao, W.-T.; Tseng, Y.-H. Gastrointestinal involvements in children with COVID-related multisystem inflammatory syndrome. Gastroenterology 2021, 160, 1887–1888. [Google Scholar] [CrossRef] [PubMed]
- Constantin, T.; Pék, T.; Horváth, Z.; Garan, D.; Szabó, A.J. Multisystem inflammatory syndrome in children (MIS-C): Implications for long COVID. Inflammopharmacology 2023, 31, 2221–2236. [Google Scholar] [CrossRef] [PubMed]
- Rowley, A.H. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat. Rev. Immunol. 2020, 20, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Wessels, P.A.; Bingler, M.A. A comparison of Kawasaki Disease and multisystem inflammatory syndrome in children. Prog. Pediatr. Cardiol. 2022, 65, 101516. [Google Scholar] [CrossRef] [PubMed]
- Sacco, K.; Castagnoli, R.; Vakkilainen, S.; Liu, C.; Delmonte, O.M.; Oguz, C.; Kaplan, I.M.; Alehashemi, S.; Burbelo, P.D.; Bhuyan, F. Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19. Nat. Med. 2022, 28, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.-L.; Wong, K.K.-Y.; Chi, S.-Q.; Zhou, A.-F.; Tang, J.-Q.; Zhou, L.-S.; Chung, P.H.-Y.; Chua, G.; Tung, K.; Wong, I. Comparative study of the clinical characteristics and epidemiological trend of 244 COVID-19 infected children with or without GI symptoms. Gut 2021, 70, 436–438. [Google Scholar] [CrossRef]
- Wells, J.M.; Brummer, R.J.; Derrien, M.; MacDonald, T.T.; Troost, F.; Cani, P.D.; Theodorou, V.; Dekker, J.; Méheust, A.; De Vos, W.M. Homeostasis of the gut barrier and potential biomarkers. Am. J. Physiol.-Gastrointest. Liver Physiol. 2017, 312, G171–G193. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef]
- Binienda, A.; Twardowska, A.; Makaro, A.; Salaga, M. Dietary carbohydrates and lipids in the pathogenesis of leaky gut syndrome: An overview. Int. J. Mol. Sci. 2020, 21, 8368. [Google Scholar] [CrossRef] [PubMed]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; He, C.; Zhu, Y.; Lu, N.-H. Role of gut microbiota on intestinal barrier function in acute pancreatitis. World J. Gastroenterol. 2020, 26, 2187. [Google Scholar] [CrossRef] [PubMed]
- Ascher, S.; Reinhardt, C. The gut microbiota: An emerging risk factor for cardiovascular and cerebrovascular disease. Eur. J. Immunol. 2018, 48, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Aron-Wisnewsky, J.; Clément, K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat. Rev. Nephrol. 2016, 12, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.W.; Kitai, T.; Hazen, S.L. Gut microbiota in cardiovascular health and disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef]
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky gut as a danger signal for autoimmune diseases. Front. Immunol. 2017, 8, 269575. [Google Scholar] [CrossRef]
- Régnier, M.; Van Hul, M.; Knauf, C.; Cani, P.D. Gut microbiome, endocrine control of gut barrier function and metabolic diseases. J. Endocrinol. 2021, 248, R67–R82. [Google Scholar] [CrossRef]
- Aziz, R.; Siles, N.; Kelley, M.; Wylie, D.; Melamed, E.; Brode, W.M. Clinical characteristics of Long COVID patients presenting to a dedicated academic post-COVID-19 clinic in Central Texas. Sci. Rep. 2023, 13, 21971. [Google Scholar] [CrossRef] [PubMed]
- Bacorn, M.; Romero-Soto, H.N.; Levy, S.; Chen, Q.; Hourigan, S.K. The Gut Microbiome of Children during the COVID-19 Pandemic. Microorganisms 2022, 10, 2460. [Google Scholar] [CrossRef] [PubMed]
- Yonker, L.M.; Gilboa, T.; Ogata, A.F.; Senussi, Y.; Lazarovits, R.; Boribong, B.P.; Bartsch, Y.C.; Loiselle, M.; Rivas, M.N.; Porritt, R.A. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.; D’Amico, F.; Brigidi, P.; Turroni, S. Gut microbiome–micronutrient interaction: The key to controlling the bioavailability of minerals and vitamins? Biofactors 2022, 48, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Corbin, K.D.; Carnero, E.A.; Dirks, B.; Igudesman, D.; Yi, F.; Marcus, A.; Davis, T.L.; Pratley, R.E.; Rittmann, B.E.; Krajmalnik-Brown, R. Host-diet-gut microbiome interactions influence human energy balance: A randomized clinical trial. Nat. Commun. 2023, 14, 3161. [Google Scholar] [CrossRef]
- Krishnan, S.; Alden, N.; Lee, K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr. Opin. Biotechnol. 2015, 36, 137–145. [Google Scholar] [CrossRef]
- Campbell, C.; Kandalgaonkar, M.R.; Golonka, R.M.; Yeoh, B.S.; Vijay-Kumar, M.; Saha, P. Crosstalk between gut microbiota and host immunity: Impact on inflammation and immunotherapy. Biomedicines 2023, 11, 294. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y. Evolution of antibody immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Giron, L.B.; Dweep, H.; Yin, X.; Wang, H.; Damra, M.; Goldman, A.R.; Gorman, N.; Palmer, C.S.; Tang, H.-Y.; Shaikh, M.W. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front. Immunol. 2021, 12, 686240. [Google Scholar]
- Trottein, F.; Sokol, H. Potential causes and consequences of gastrointestinal disorders during a SARS-CoV-2 infection. Cell Rep. 2020, 32. [Google Scholar] [CrossRef] [PubMed]
- Ancona, G.; Alagna, L.; Alteri, C.; Palomba, E.; Tonizzo, A.; Pastena, A.; Muscatello, A.; Gori, A.; Bandera, A. Gut and airway microbiota dysbiosis and their role in COVID-19 and long-COVID. Front. Immunol. 2023, 14, 1080043. [Google Scholar] [CrossRef]
- Komaroff, A.L.; Lipkin, W.I. ME/CFS and Long COVID share similar symptoms and biological abnormalities: Road map to the literature. Front. Med. 2023, 10, 1187163. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.-Y.; Ng, S.S.S.; Zhang, F.; Li, A.Y.; Lu, W.; Hui, D.S.-C. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Yi, B.; Wu, J.; Lu, J. The microbiome in post-acute infection syndrome (PAIS). Comput. Struct. Biotechnol. J. 2023. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.M.; Singer, D.; Makrides, V.; Huggel, K.; Pos, K.M.; Wagner, C.A.; Kuba, K.; Danilczyk, U.; Skovby, F.; Kleta, R. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology 2009, 136, 872–882.e873. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Perlot, T.; Penninger, J.M. ACE2–From the renin–angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013, 15, 866–873. [Google Scholar] [CrossRef]
- Viana, S.D.; Nunes, S.; Reis, F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities–role of gut microbiota dysbiosis. Ageing Res. Rev. 2020, 62, 101123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, Y.; Ma, Y.; Chen, P.; Tang, J.; Yang, B.; Li, H.; Liang, M.; Xue, Y.; Liu, Y. Gut microbiota dysbiosis correlates with long COVID-19 at one-year after discharge. J. Korean Med. Sci. 2023, 38. [Google Scholar] [CrossRef]
- Suskun, C.; Kilic, O.; Yilmaz Ciftdogan, D.; Guven, S.; Karbuz, A.; Ozkaya Parlakay, A.; Kara, Y.; Kacmaz, E.; Sahin, A.; Boga, A. Intestinal microbiota composition of children with infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and multisystem inflammatory syndrome (MIS-C). Eur. J. Pediatr. 2022, 181, 3175–3191. [Google Scholar] [CrossRef]
- Romani, L.; Del Chierico, F.; Macari, G.; Pane, S.; Ristori, M.V.; Guarrasi, V.; Gardini, S.; Pascucci, G.R.; Cotugno, N.; Perno, C.F. The relationship between pediatric gut microbiota and SARS-CoV-2 infection. Front. Cell. Infect. Microbiol. 2022, 12, 908492. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, B.; Luckey, D.; Taneja, V. Autoimmunity-associated gut commensals modulate gut permeability and immunity in humanized mice. Mil. Med. 2019, 184, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef]
- Santaolalla, R.; Abreu, M.T. Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 2012, 28, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.; Trinchieri, G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat. Rev. Immunol. 2011, 11, 9–20. [Google Scholar] [CrossRef]
- Yonker, L.; Neilan, A.; Bartsch, Y. Pediatric SARS-CoV-2: Clinical presentation, infectivity, and immune responses [manuscript published online ahead of print 20 August 2020]. J Pediatr 2020, 10. [Google Scholar]
- Wang, Y.; Zhang, L.; Sang, L.; Ye, F.; Ruan, S.; Zhong, B.; Song, T.; Alshukairi, A.N.; Chen, R.; Zhang, Z. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Investig. 2020, 130, 5235–5244. [Google Scholar] [CrossRef] [PubMed]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Minami, S.; Matsumoto, N.; Omori, H.; Nakamura, Y.; Tamiya, S.; Nouda, R.; Nurdin, J.A.; Yamasaki, M.; Kotaki, T.; Kanai, Y. Effective SARS-CoV-2 replication of monolayers of intestinal epithelial cells differentiated from human induced pluripotent stem cells. Sci. Rep. 2023, 13, 11610. [Google Scholar] [CrossRef]
- Zang, R.; Castro, M.F.G.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef] [PubMed]
- Stanifer, M.L.; Kee, C.; Cortese, M.; Zumaran, C.M.; Triana, S.; Mukenhirn, M.; Kraeusslich, H.-G.; Alexandrov, T.; Bartenschlager, R.; Boulant, S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020, 32. [Google Scholar] [CrossRef]
- Fasano, A.; Not, T.; Wang, W.; Uzzau, S.; Berti, I.; Tommasini, A.; Goldblum, S.E. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 2000, 355, 1518–1519. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. Regulation of intercellular tight junctions by zonula occludens toxin and its eukaryotic analogue zonulin. Ann. N. Y. Acad. Sci. 2000, 915, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Uzzau, S.; Goldblum, S.E.; Fasano, A. Human zonulin, a potential modulator of intestinal tight junctions. J. Cell Sci. 2000, 113, 4435–4440. [Google Scholar] [CrossRef] [PubMed]
- Kılıç, A.O.; Akın, F.; Yazar, A.; Metin Akcan, Ö.; Topcu, C.; Aydın, O. Zonulin and claudin-5 levels in multisystem inflammatory syndrome and SARS-CoV-2 infection in children. J. Paediatr. Child Health 2022, 58, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Okuyucu, M.; Kehribar, D.Y.; Çapraz, M.; Çapraz, A.; Arslan, M.; Çelik, Z.B.; Usta, B.; Birinci, A.; Ozgen, M.; Özgen, M. The relationship between COVID-19 disease severity and zonulin levels. Cureus 2022, 14. [Google Scholar] [CrossRef]
- Hensley-McBain, T.; Manuzak, J.A. Zonulin as a biomarker and potential therapeutic target in multisystem inflammatory syndrome in children. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Palomino-Kobayashi, L.A.; Ymaña, B.; Ruiz, J.; Mayanga-Herrera, A.; Ugarte-Gil, M.F.; Pons, M.J. Zonulin, a marker of gut permeability, is associated with mortality in a cohort of hospitalised peruvian COVID-19 patients. Front. Cell. Infect. Microbiol. 2022, 12, 1000291. [Google Scholar]
- Llorens, S.; Nava, E.; Muñoz-López, M.; Sánchez-Larsen, Á.; Segura, T. Neurological symptoms of COVID-19: The zonulin hypothesis. Front. Immunol. 2021, 12, 665300. [Google Scholar] [CrossRef]
- Drago, S.; El Asmar, R.; Di Pierro, M.; Grazia Clemente, M.; Sapone, A.T.A.; Thakar, M.; Iacono, G.; Carroccio, A.; D’Agate, C.; Not, T. Gliadin, zonulin and gut permeability: Effects on celiac and non-celiac intestinal mucosa and intestinal cell lines. Scand. J. Gastroenterol. 2006, 41, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, J.; Vogelsang, H.; Hübl, W.; Waldhoer, T.; Lochs, H. Intestinal permeability and the prediction of relapse in Crohn’s disease. Lancet 1993, 341, 1437–1439. [Google Scholar] [CrossRef] [PubMed]
- Rivas, M.N.; Wakita, D.; Franklin, M.K.; Carvalho, T.T.; Abolhesn, A.; Gomez, A.C.; Fishbein, M.C.; Chen, S.; Lehman, T.J.; Sato, K. Intestinal permeability and IgA provoke immune vasculitis linked to cardiovascular inflammation. Immunity 2019, 51, 508–521.e506. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. All disease begins in the (leaky) gut: Role of zonulin-mediated gut permeability in the pathogenesis of some chronic inflammatory diseases. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Brogna, C.; Cristoni, S.; Petrillo, M.; Querci, M.; Piazza, O.; Van den Eede, G. Toxin-like peptides in plasma, urine and faecal samples from COVID-19 patients. F1000Research 2021, 10. [Google Scholar] [CrossRef]
- Li, C.; Gao, M.; Zhang, W.; Chen, C.; Zhou, F.; Hu, Z.; Zeng, C. Zonulin regulates intestinal permeability and facilitates enteric bacteria permeation in coronary artery disease. Sci. Rep. 2016, 6, 29142. [Google Scholar] [CrossRef] [PubMed]
- Ghazizadeh Esslami, G.; Mamishi, S.; Pourakbari, B.; Mahmoudi, S. Systematic review and meta-analysis on the serological, immunological, and cardiac parameters of the multisystem inflammatory syndrome (MIS-C) associated with SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28927. [Google Scholar] [CrossRef] [PubMed]
- El Asmar, R.; Panigrahi, P.; Bamford, P.; Berti, I.; Not, T.; Coppa, G.V.; Catassi, C.; Fasano, A. Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterology 2002, 123, 1607–1615. [Google Scholar] [CrossRef]
- Thomas, K.E.; Sapone, A.; Fasano, A.; Vogel, S.N. Gliadin stimulation of murine macrophage inflammatory gene expression and intestinal permeability are MyD88-dependent: Role of the innate immune response in Celiac disease. J. Immunol. 2006, 176, 2512–2521. [Google Scholar] [CrossRef]
- Fasano, A. Intestinal permeability and its regulation by zonulin: Diagnostic and therapeutic implications. Clin. Gastroenterol. Hepatol. 2012, 10, 1096–1100. [Google Scholar] [CrossRef]
- Ogata, A.F.; Maley, A.M.; Wu, C.; Gilboa, T.; Norman, M.; Lazarovits, R.; Mao, C.-P.; Newton, G.; Chang, M.; Nguyen, K. Ultra-sensitive serial profiling of SARS-CoV-2 antigens and antibodies in plasma to understand disease progression in COVID-19 patients with severe disease. Clin. Chem. 2020, 66, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Kikuta, J.; Matsui, T.; Hasegawa, T.; Fujii, K.; Okuzaki, D.; Liu, Y.-c.; Yoshioka, T.; Seno, S.; Motooka, D. Periportal macrophages protect against commensal-driven liver inflammation. Nature 2024, 1–9. [Google Scholar] [CrossRef]
- Chai, X.; Hu, L.; Zhang, Y.; Han, W.; Lu, Z.; Ke, A.; Zhou, J.; Shi, G.; Fang, N.; Fan, J. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv 2002. [Google Scholar] [CrossRef]
- Luxenburger, H.; Thimme, R. SARS-CoV-2 and the liver: Clinical and immunological features in chronic liver disease. Gut 2023, 72, 1783–1794. [Google Scholar] [CrossRef]
- Lücke, J.; Nawrocki, M.; Schnell, J.; Meins, N.; Heinrich, F.; Zhang, T.; Bertram, F.; Sabihi, M.; Böttcher, M.; Blankenburg, T. TNFα aggravates detrimental effects of SARS-CoV-2 infection in the liver. Front. Immunol. 2023, 14, 1151937. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Ballester, M.P.; Soffientini, U.; Jalan, R.; Mehta, G. SARS-CoV-2 infection and liver involvement. Hepatol. Int. 2022, 16, 755–774. [Google Scholar] [CrossRef] [PubMed]
- Lazova, S.; Alexandrova, T.; Gorelyova-Stefanova, N.; Atanasov, K.; Tzotcheva, I.; Velikova, T. Liver involvement in children with COVID-19 and multisystem inflammatory syndrome: A single-center Bulgarian observational study. Microorganisms 2021, 9, 1958. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-H.; Zheng, K.I.; Targher, G.; Byrne, C.D.; Zheng, M.-H. Abnormal liver enzymes in children and infants with COVID-19: A narrative review of case-series studies. Pediatr. Obes. 2020, 15, e12723. [Google Scholar] [CrossRef]
- Yao, N.; Wang, S.; Lian, J.; Sun, Y.; Zhang, G.; Kang, W.; Kang, W. Clinical characteristics and influencing factors of patients with novel coronavirus pneumonia combined with liver injury in Shaanxi region. Zhonghua Gan Zang Bing Za Zhi = Zhonghua Ganzangbing Zazhi = Chin. J. Hepatol. 2020, 28, 234–239. [Google Scholar]
- Brogna, C.; Brogna, B.; Bisaccia, D.R.; Lauritano, F.; Marino, G.; Montano, L.; Cristoni, S.; Prisco, M.; Piscopo, M. Could SARS-CoV-2 have bacteriophage behavior or induce the activity of other bacteriophages? Vaccines 2022, 10, 708. [Google Scholar] [CrossRef]
- Brogna, C.; Viduto, V.; Fabrowski, M.; Cristoni, S.; Marino, G.; Montano, L.; Piscopo, M. The importance of the gut microbiome in the pathogenesis and transmission of SARS-CoV-2: Someone on Earth: “.. we moved at the speed of Science!”-Science from the center of the Universe: “Hey man, I’m still waiting for you in the 50s!”. Gut Microbes 2023, 15, 2244718. [Google Scholar] [CrossRef]
- Petrillo, M.; Querci, M.; Brogna, C.; Ponti, J.; Cristoni, S.; Markov, P.V.; Valsesia, A.; Leoni, G.; Benedetti, A.; Wiss, T. Evidence of SARS-CoV-2 bacteriophage potential in human gut microbiota. F1000Research 2022, 11, 292. [Google Scholar] [CrossRef]
- Brogna, C.; Costanzo, V.; Brogna, B.; Bisaccia, D.R.; Brogna, G.; Giuliano, M.; Montano, L.; Viduto, V.; Cristoni, S.; Fabrowski, M. Analysis of bacteriophage behavior of a human RNA virus, SARS-CoV-2, through the integrated approach of immunofluorescence microscopy, proteomics and D-amino acid quantification. Int. J. Mol. Sci. 2023, 24, 3929. [Google Scholar] [CrossRef]
- Brogna, C.; Cristoni, S.; Petrillo, M.; Bisaccia, D.R.; Lauritano, F.; Montano, L.; Prisco, M.; Piscopo, M. The first report on detecting SARS-CoV-2 inside bacteria of the human gut microbiome: A case series on asymptomatic family members and a child with COVID-19. F1000Research 2022, 11, 135. [Google Scholar] [CrossRef]
- Petrillo, M.; Brogna, C.; Cristoni, S.; Querci, M.; Piazza, O.; Van den Eede, G. Increase of SARS-CoV-2 RNA load in faecal samples prompts for rethinking of SARS-CoV-2 biology and COVID-19 epidemiology. F1000Research 2021, 10. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.; Yeoh, Y.K.; Li, A.Y.; Zhan, H.; Wan, Y.; Chung, A.C.; Cheung, C.P.; Chen, N. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020, 159, 944–955.e948. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Noval Rivas, M.; Porritt, R.A.; Cheng, M.H.; Bahar, I.; Arditi, M. Multisystem inflammatory syndrome in children and long COVID: The SARS-CoV-2 viral superantigen hypothesis. Front. Immunol. 2022, 13, 941009. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Cheng, M.H.; Zhang, S.; Porritt, R.A.; Noval Rivas, M.; Paschold, L.; Willscher, E.; Binder, M.; Arditi, M.; Bahar, I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. 2020, 117, 25254–25262. [Google Scholar] [CrossRef]
- Amormino, C.; Tedeschi, V.; Paldino, G.; Arcieri, S.; Fiorillo, M.T.; Paiardini, A.; Tuosto, L.; Kunkl, M. SARS-CoV-2 spike does not possess intrinsic superantigen-like inflammatory activity. Cells 2022, 11, 2526. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.C. MIS-C: Myths have been debunked, but mysteries remain. Nat. Rev. Rheumatol. 2023, 19, 70–71. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.-E.; Song, J.; Grifoni, A.; Shimizu, C.; Tremoulet, A.H.; Dummer, K.B.; Burns, J.C.; Sette, A.; Franco, A. T cells in multisystem inflammatory syndrome in children (MIS-C) have a predominant CD4+ T helper response to SARS-CoV-2 peptides and numerous virus-specific CD4− CD8− double-negative T cells. Int. J. Mol. Sci. 2022, 23, 7219. [Google Scholar] [CrossRef]
- Sigal, G.B.; Novak, T.; Mathew, A.; Chou, J.; Zhang, Y.; Manjula, N.; Bathala, P.; Joe, J.; Padmanabhan, N.; Romero, D. Measurement of severe acute respiratory syndrome coronavirus 2 antigens in plasma of pediatric patients with acute coronavirus disease 2019 or multisystem inflammatory syndrome in children using an ultrasensitive and quantitative immunoassay. Clin. Infect. Dis. 2022, 75, 1351–1358. [Google Scholar] [CrossRef]
- Levy, N.; Koppel, J.H.; Kaplan, O.; Yechiam, H.; Shahar-Nissan, K.; Cohen, N.K.; Shavit, I. Severity and incidence of multisystem inflammatory syndrome in children during 3 SARS-CoV-2 pandemic waves in Israel. Jama 2022, 327, 2452–2454. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Toba, S.; Itakura, Y.; Chambaro, H.M.; Kishimoto, M.; Tabata, K.; Intaruck, K.; Uemura, K.; Sanaki, T.; Sato, A. SARS-CoV-2 bearing a mutation at the S1/S2 cleavage site exhibits attenuated virulence and confers protective immunity. Mbio 2021, 12. [Google Scholar] [CrossRef]
- Zhang, L.; Mann, M.; Syed, Z.A.; Reynolds, H.M.; Tian, E.; Samara, N.L.; Zeldin, D.C.; Tabak, L.A.; Ten Hagen, K.G. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. USA 2021, 118, e2109905118. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2 intermittent virulence as a result of natural selection. COVID 2022, 2, 1089–1101. [Google Scholar] [CrossRef]
- Coomes, E.A.; Haghbayan, H. Interleukin-6 in COVID-19: A systematic review and meta-analysis. Rev. Med. Virol. 2020, 30, 1–9. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, J.; Yang, Y.; Ma, H.; Li, Z.; Zhang, J.; Cheng, J.; Zhang, X.; Zhao, Y.; Xia, Z. The role of interleukin-6 in monitoring severe case of coronavirus disease 2019. EMBO Mol. Med. 2020, 12, e12421. [Google Scholar] [CrossRef]
- Zhu, J.; Pang, J.; Ji, P.; Zhong, Z.; Li, H.; Li, B.; Zhang, J. Elevated interleukin-6 is associated with severity of COVID-19: A meta-analysis. J. Med. Virol. 2021, 93, 35. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.-X.; Agbana, Y.L.; Sun, Z.-S.; Fei, S.-W.; Zhao, H.-Q.; Zhou, X.-N.; Chen, J.-H.; Kassegne, K. Increased interleukin-6 is associated with long COVID-19: A systematic review and meta-analysis. Infect. Dis. Poverty 2023, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Fatima, R.; Assaly, R. Elevated interleukin-6 and severe COVID-19: A meta-analysis. J. Med. Virol. 2020, 92, 2283. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, Y.C.; Wang, C.; Zohar, T.; Fischinger, S.; Atyeo, C.; Burke, J.S.; Kang, J.; Edlow, A.G.; Fasano, A.; Baden, L.R. Humoral signatures of protective and pathological SARS-CoV-2 infection in children. Nat. Med. 2021, 27, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Karwaciak, I.; Sałkowska, A.; Karaś, K.; Dastych, J.; Ratajewski, M. Nucleocapsid and spike proteins of the coronavirus SARS-CoV-2 induce il6 in monocytes and macrophages—Potential implications for cytokine storm syndrome. Vaccines 2021, 9, 54. [Google Scholar] [CrossRef]
- Hasan, A.; Rahim, R.; Nakayama, E.E.; Uno, K.; Hasan, N.; Rahman, M.; Shioda, T. Enhancement of IL-6 production induced by SARS-CoV-2 Nucleocapsid protein and Bangladeshi COVID-19 patients’ sera. Viruses 2023, 15, 2018. [Google Scholar] [CrossRef]
- Aliyu, M.; Zohora, F.T.; Anka, A.U.; Ali, K.; Maleknia, S.; Saffarioun, M.; Azizi, G. Interleukin-6 cytokine: An overview of the immune regulation, immune dysregulation, and therapeutic approach. Int. Immunopharmacol. 2022, 111, 109130. [Google Scholar] [CrossRef]
- Kishimoto, T. The biology of interleukin-6. Blood 1989, 74, 1–10. [Google Scholar] [CrossRef]
- Chomarat, P.; Banchereau, J.; Davoust, J.; Karolina Palucka, A. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat. Immunol. 2000, 1, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Narimatsu, M.; Maeda, H.; Itoh, S.; Atsumi, T.; Ohtani, T.; Nishida, K.; Itoh, M.; Kamimura, D.; Park, S.-J.; Mizuno, K. Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells. Mol. Cell. Biol. 2001. [Google Scholar] [CrossRef]
- Teague, T.K.; Schaefer, B.C.; Hildeman, D.; Bender, J.; Mitchell, T.; Kappler, J.W.; Marrack, P. Activation-induced inhibition of interleukin 6–mediated T cell survival and signal transducer and activator of transcription 1 signaling. J. Exp. Med. 2000, 191, 915–926. [Google Scholar] [CrossRef]
- Curnow, S.J.; Scheel-Toellner, D.; Jenkinson, W.; Raza, K.; Durrani, O.M.; Faint, J.M.; Rauz, S.; Wloka, K.; Pilling, D.; Rose-John, S. Inhibition of T cell apoptosis in the aqueous humor of patients with uveitis by IL-6/soluble IL-6 receptor trans-signaling. J. Immunol. 2004, 173, 5290–5297. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Kishimoto, T. IL-6: Regulator of Treg/Th17 balance. Eur. J. Immunol. 2010, 40, 1830–1835. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. Therapeutic targeting of the interleukin-6 receptor. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 199–219. [Google Scholar] [CrossRef]
- Neurath, M.F.; Finotto, S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011, 22, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Aricha, R.; Mizrachi, K.; Fuchs, S.; Souroujon, M.C. Blocking of IL-6 suppresses experimental autoimmune myasthenia gravis. J. Autoimmun. 2011, 36, 135–141. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E.; Maerz, M.D.; Buckner, J.H. IL-6: A cytokine at the crossroads of autoimmunity. Curr. Opin. Immunol. 2018, 55, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Iwakura, T.; Matsui, K.; Kawaguchi, H.; Obana, M.; Hayama, A.; Maeda, M.; Izumi, Y.; Komuro, I.; Ohsugi, Y. IL-6-mediated Th17 differentiation through RORγt is essential for the initiation of experimental autoimmune myocarditis. Cardiovasc. Res. 2011, 91, 640–648. [Google Scholar] [CrossRef]
- Narazaki, M.; Tanaka, T.; Kishimoto, T. The role and therapeutic targeting of IL-6 in rheumatoid arthritis. Expert Rev. Clin. Immunol. 2017, 13, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, K.A.; Manieri, N.A.; Liu, T.-C.; Stappenbeck, T.S. IL-6 stimulates intestinal epithelial proliferation and repair after injury. PLoS ONE 2014, 9, e114195. [Google Scholar] [CrossRef]
- Kawashima, T.; Ikari, N.; Kouchi, T.; Kowatari, Y.; Kubota, Y.; Shimojo, N.; Tsuji, N.M. The molecular mechanism for activating IgA production by Pediococcus acidilactici K15 and the clinical impact in a randomized trial. Sci. Rep. 2018, 8, 5065. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, B.; Wang, T.; Gao, L.; Yang, Z.-j.; Wang, F.-f.; Shang, H.-w.; Hua, R.; Xu, J.-d. Biological characteristics of IL-6 and related intestinal diseases. Int. J. Biol. Sci. 2021, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, Y.; Cui, T.; Zhang, J. IL-6/STAT3 signaling pathway regulates the proliferation and damage of intestinal epithelial cells in patients with ulcerative colitis via H3K27ac. Exp. Ther. Med. 2021, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Isaza-Correa, J.; Ryan, L.; Kelly, L.; Allen, J.; Melo, A.; Jones, J.; Huggard, D.; Ryan, E.; Ó Maoldomhnaigh, C.; Geoghehan, S. Innate immune dysregulation in multisystem inflammatory syndrome in children (MIS-C). Sci. Rep. 2023, 13, 16463. [Google Scholar] [CrossRef] [PubMed]
- Rybkina, K.; Bell, J.N.; Bradley, M.C.; Wohlbold, T.; Scafuro, M.; Meng, W.; Korenberg, R.C.; Davis-Porada, J.; Anderson, B.R.; Weller, R.J. SARS-CoV-2 infection and recovery in children: Distinct T cell responses in MIS-C compared to COVID-19. J. Exp. Med. 2023, 220, e20221518. [Google Scholar] [CrossRef] [PubMed]
- Gruber, C.N.; Patel, R.S.; Trachtman, R.; Lepow, L.; Amanat, F.; Krammer, F.; Wilson, K.M.; Onel, K.; Geanon, D.; Tuballes, K. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C). Cell 2023, 186, 3325. [Google Scholar] [CrossRef] [PubMed]
- Zerra, P.E.; Stowell, J.; Verkerke, H.; McCoy, J.; Jones, J.; Graciaa, S.; Lu, A.; Hussaini, L.; Anderson, E.J.; Rostad, C.A. Factor H autoantibodies contribute to complement dysregulation in multisystem inflammatory syndrome in children (MIS-C). Am. J. Hematol. 2023, 98, E98–E101. [Google Scholar] [CrossRef]
- Lee, M.J.; Leong, M.W.; Rustagi, A.; Beck, A.; Zeng, L.; Holmes, S.; Qi, L.S.; Blish, C.A. SARS-CoV-2 escapes direct NK cell killing through Nsp1-mediated downregulation of ligands for NKG2D. Cell Rep. 2022, 41. [Google Scholar] [CrossRef]
- Brogna, C.; Montano, L.; Zanolin, M.E.; Bisaccia, D.R.; Ciammetti, G.; Viduto, V.; Fabrowski, M.; Baig, A.M.; Gerlach, J.; Gennaro, I. A retrospective cohort study on early antibiotic use in vaccinated and unvaccinated COVID-19 patients. J. Med. Virol. 2024, 96, e29507. [Google Scholar] [CrossRef] [PubMed]
- Elvan-Tuz, A.; Ekemen-Keles, Y.; Karadag-Oncel, E.; Yilmaz, D.; Ozcifci, G.; Durak, F. Nowadays a new MIS-C mimicker: Group A streptococcal infections. Pediatr. Infect. Dis. J. 2023, 42, e129. [Google Scholar] [CrossRef] [PubMed]
- Della-Torre, E.; Criscuolo, E.; Lanzillotta, M.; Locatelli, M.; Clementi, N.; Mancini, N.; Dagna, L.; Group, C.-B.S. IL-1 and IL-6 inhibition affects the neutralising activity of anti-SARS-CoV-2 antibodies in patients with COVID-19. Lancet Rheumatol. 2021, 3, e829. [Google Scholar] [CrossRef] [PubMed]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Charness, M.E.; Gupta, K.; Stack, G.; Strymish, J.; Adams, E.; Lindy, D.C.; Mohri, H.; Ho, D.D. Rebound of SARS-CoV-2 infection after nirmatrelvir–ritonavir treatment. N. Engl. J. Med. 2022, 387, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, V.; Di Lucia, P.; Ravà, M.; Marotta, D.; Bono, E.; Grassi, S.; Donnici, L.; Cannalire, R.; Stefanelli, I.; Ferraro, A. Nirmatrelvir treatment of SARS-CoV-2-infected mice blunts antiviral adaptive immune responses. EMBO Mol. Med. 2023, 15, e17580. [Google Scholar] [CrossRef]
- Veres-Székely, A.; Szász, C.; Pap, D.; Szebeni, B.; Bokrossy, P.; Vannay, Á. Zonulin as a potential therapeutic target in microbiota-gut-brain axis disorders: Encouraging results and emerging questions. Int. J. Mol. Sci. 2023, 24, 7548. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, Z.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb. Cell Factories 2020, 19, 1–11. [Google Scholar] [CrossRef]
- Gupta, P.; Andrew, H.; Kirschner, B.S.; Guandalini, S. Is Lactobacillus GG helpful in children with Crohn’s disease? Results of a preliminary, open-label study. J. Pediatr. Gastroenterol. Nutr. 2000, 31, 453–457. [Google Scholar]
- Ewaschuk, J.B.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Looijer-van Langen, M.; Madsen, K.L. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol.-Gastrointest. Liver Physiol. 2008, 295, G1025–G1034. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, Z.; Tang, P.; Wu, Y.; Zhang, A.; Li, D.; Wang, C.-Z.; Wan, J.-Y.; Yao, H.; Yuan, C.-S. Probiotics fortify intestinal barrier function: A systematic review and meta-analysis of randomized trials. Front. Immunol. 2023, 14, 1143548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, X.; Jiang, Y.; Zhao, W.; Guo, T.; Cao, Y.; Teng, J.; Hao, X.; Zhao, J.; Yang, Z. Antioxidant status and gut microbiota change in an aging mouse model as influenced by exopolysaccharide produced by Lactobacillus plantarum YW11 isolated from Tibetan kefir. J. Dairy Sci. 2017, 100, 6025–6041. [Google Scholar] [CrossRef] [PubMed]
- Petrof, E.O.; Kojima, K.; Ropeleski, M.J.; Musch, M.W.; Tao, Y.; De Simone, C.; Chang, E.B. Probiotics inhibit nuclear factor-κB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology 2004, 127, 1474–1487. [Google Scholar] [CrossRef]
- Martorell, P.; Alvarez, B.; Llopis, S.; Navarro, V.; Ortiz, P.; Gonzalez, N.; Balaguer, F.; Rojas, A.; Chenoll, E.; Ramon, D. Heat-treated Bifidobacterium longum CECT-7347: A whole-cell postbiotic with antioxidant, anti-inflammatory, and gut-barrier protection properties. Antioxidants 2021, 10, 536. [Google Scholar] [CrossRef]
- Mullish, B.H.; Marchesi, J.R.; McDonald, J.A.; Pass, D.A.; Masetti, G.; Michael, D.R.; Plummer, S.; Jack, A.A.; Davies, T.S.; Hughes, T.R. Probiotics reduce self-reported symptoms of upper respiratory tract infection in overweight and obese adults: Should we be considering probiotics during viral pandemics? Gut Microbes 2021, 13, 1900997. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Borrazzo, C.; Pinacchio, C.; Santinelli, L.; Innocenti, G.P.; Cavallari, E.N.; Celani, L.; Marazzato, M.; Alessandri, F.; Ruberto, F. Oral bacteriotherapy in patients with COVID-19: A retrospective cohort study. Front. Nutr. 2021, 7, 613928. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.; Davies, T.; Jack, A.; Masetti, G.; Marchesi, J.; Wang, D.; Mullish, B.; Plummer, S. Daily supplementation with the Lab4P probiotic consortium induces significant weight loss in overweight adults. Sci. Rep. 2021, 11, 5. [Google Scholar] [CrossRef]
- García-Mena, J.; Corona-Cervantes, K.; Cuervo-Zanatta, D.; Benitez-Guerrero, T.; Vélez-Ixta, J.M.; Zavala-Torres, N.G.; Villalobos-Flores, L.E.; Hernández-Quiroz, F.; Perez-Cruz, C.; Murugesan, S. Gut microbiota in a population highly affected by obesity and type 2 diabetes and susceptibility to COVID-19. World J. Gastroenterol. 2021, 27, 7065. [Google Scholar] [CrossRef]
- Ivashkin, V.; Fomin, V.; Moiseev, S.; Brovko, M.; Maslennikov, R.; Ulyanin, A.; Sholomova, V.; Vasilyeva, M.; Trush, E.; Shifrin, O. Efficacy of a Probiotic Consisting of Lacticaseibacillus rhamnosus PDV 1705, Bifidobacterium bifidum PDV 0903, Bifidobacterium longum subsp. infantis PDV 1911, and Bifidobacterium longum subsp. longum PDV 2301 in the Treatment of Hospitalized Patients with COVID-19: A Randomized Controlled Trial. Probiotics Antimicrob. Proteins 2021, 1–19. [Google Scholar]
- Kurian, S.J.; Unnikrishnan, M.K.; Miraj, S.S.; Bagchi, D.; Banerjee, M.; Reddy, B.S.; Rodrigues, G.S.; Manu, M.K.; Saravu, K.; Mukhopadhyay, C. Probiotics in prevention and treatment of COVID-19: Current perspective and future prospects. Arch. Med. Res. 2021, 52, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, H.S.; Bilen, Ö. Oral booster probiotic bifidobacteria in SARS-COV-2 patients. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211059677. [Google Scholar] [CrossRef]
- Gutiérrez-Castrellón, P.; Gandara-Martí, T.; Abreu Y Abreu, A.T.; Nieto-Rufino, C.D.; López-Orduña, E.; Jiménez-Escobar, I.; Jiménez-Gutiérrez, C.; López-Velazquez, G.; Espadaler-Mazo, J. Probiotic improves symptomatic and viral clearance in Covid19 outpatients: A randomized, quadruple-blinded, placebo-controlled trial. Gut Microbes 2022, 14, 2018899. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Z.; Mak, J.W.; Chow, K.M.; Lui, G.; Li, T.C.; Wong, C.K.; Chan, P.K.; Ching, J.Y.; Fujiwara, Y. Gut microbiota-derived synbiotic formula (SIM01) as a novel adjuvant therapy for COVID-19: An open-label pilot study. J. Gastroenterol. Hepatol. 2022, 37, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Saviano, A.; Potenza, A.; Siciliano, V.; Petruzziello, C.; Tarli, C.; Migneco, A.; Nasella, F.; Franceschi, F.; Ojetti, V. COVID-19 pneumonia and gut inflammation: The role of a mix of three probiotic strains in reducing inflammatory markers and need for oxygen support. J. Clin. Med. 2022, 11, 3758. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, B.R.; Hao, Q. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst. Rev. 2022. [Google Scholar]
- Alenazy, M.F.; Aljohar, H.I.; Alruwaili, A.R.; Daghestani, M.H.; Alonazi, M.A.; Labban, R.S.; El-Ansary, A.K.; Balto, H.A. Gut microbiota dynamics in relation to long-COVID-19 syndrome: Role of probiotics to combat psychiatric complications. Metabolites 2022, 12, 912. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Capozzi, V.; Russo, P.; Drider, D.; Spano, G.; Fiocco, D. Immunobiosis and probiosis: Antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl. Microbiol. Biotechnol. 2018, 102, 9949–9958. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, D.K.; Ha, N.J.; Shin, H.S. Antiviral effects of Lactobacillus ruminis SPM0211 and Bifidobacterium longum SPM1205 and SPM1206 on rotavirus-infected Caco-2 cells and a neonatal mouse model. J. Microbiol. 2015, 53, 796–803. [Google Scholar] [CrossRef]
- Shi, H.; Zuo, Y.; Navaz, S.; Harbaugh, A.; Hoy, C.K.; Gandhi, A.A.; Sule, G.; Yalavarthi, S.; Gockman, K.; Madison, J.A. Endothelial cell–activating antibodies in COVID-19. Arthritis Rheumatol. 2022, 74, 1132–1138. [Google Scholar] [CrossRef]
- Blagova, O.; Varionchik, N.; Zaidenov, V.; Savina, P.; Sarkisova, N. Anti-heart antibodies levels and their correlation with clinical symptoms and outcomes in patients with confirmed or suspected diagnosis COVID-19. Eur. J. Immunol. 2021, 51, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Buchrieser, J.; Dufloo, J.; Hubert, M.; Monel, B.; Planas, D.; Rajah, M.M.; Planchais, C.; Porrot, F.; Guivel-Benhassine, F.; Van der Werf, S. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2021, 40, e107405. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Naseem, A.; Siddiqui, Z.I. SARS-CoV-2 Syncytium under the radar: Molecular insights of the spike-induced syncytia and potential strategies to limit SARS-CoV-2 replication. J. Clin. Med. 2023, 12, 6079. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).