A Serological Multiplexed Immunoassay (MIA) Detects Antibody Reactivity to SARS-CoV-2 and Other Viral Pathogens in Liberia and Is Configurable as a Multiplexed Inhibition Test (MINT)
Abstract
1. Introduction
2. Materials and Methods
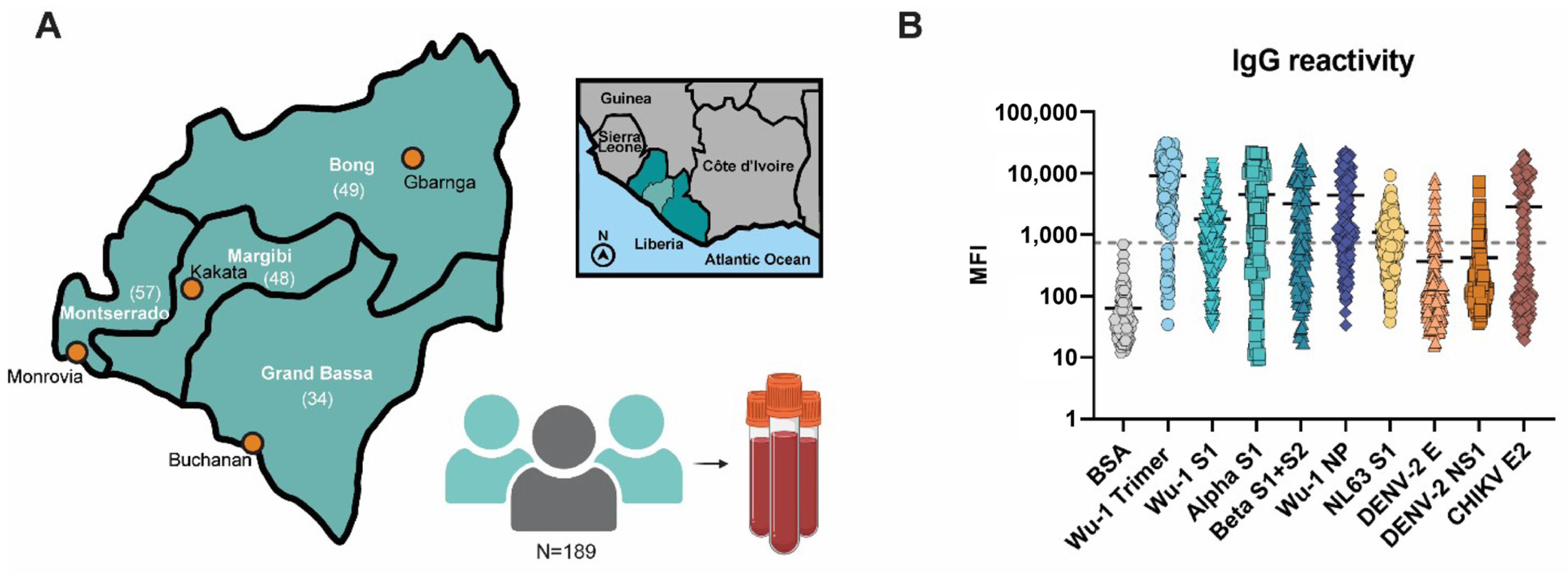
2.1. Sample Collection and Transportation
2.2. Protein Expression and Purification
2.2.1. Human ACE-2 (hACE-2)
2.2.2. SARS-CoV-2 S Trimer
2.2.3. SARS-CoV-2 NP
2.2.4. CHIKV E2
2.3. Coupling of Microspheres with Recombinant Antigens
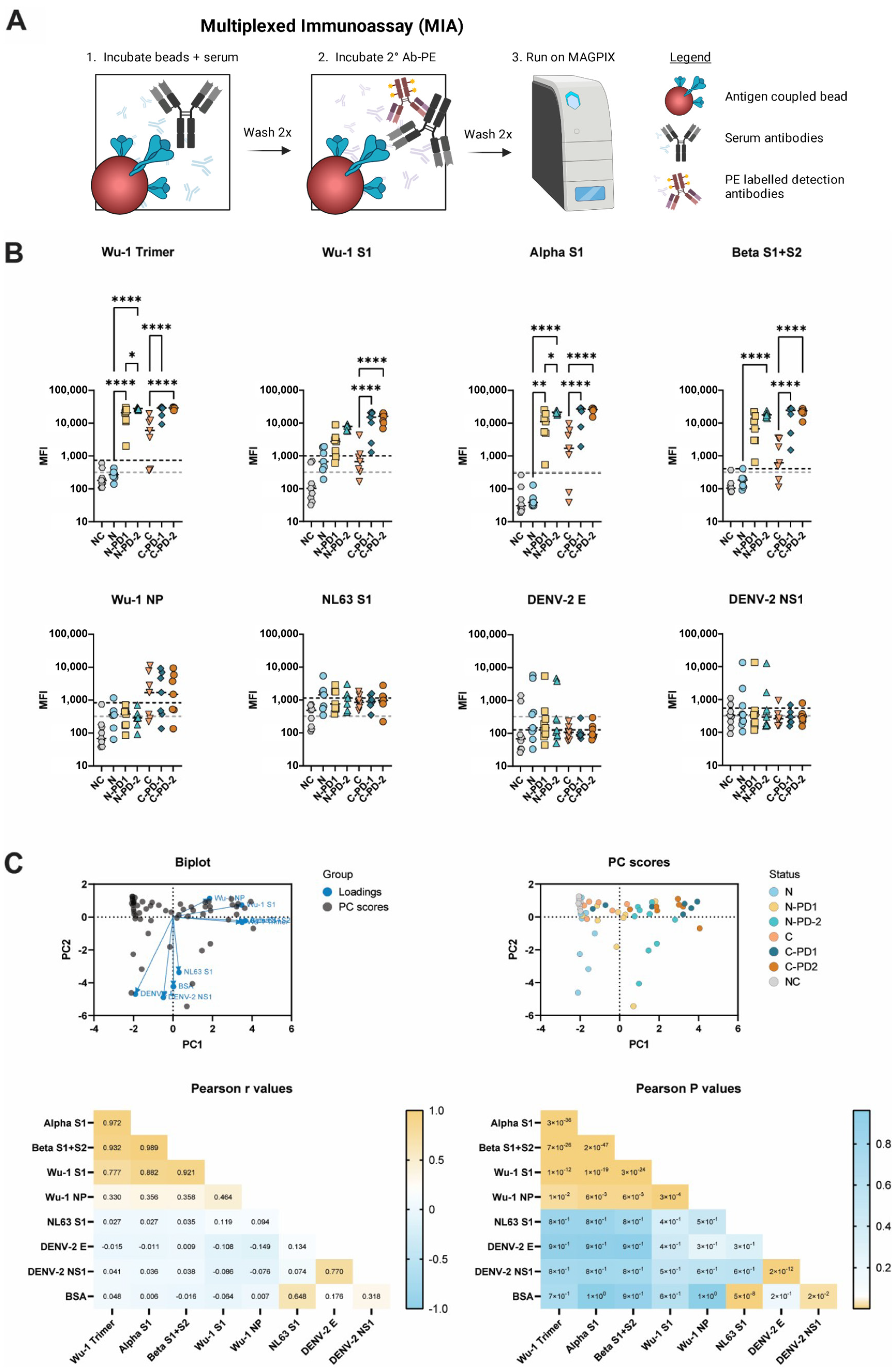
2.4. Microsphere Immunoassay (MIA)
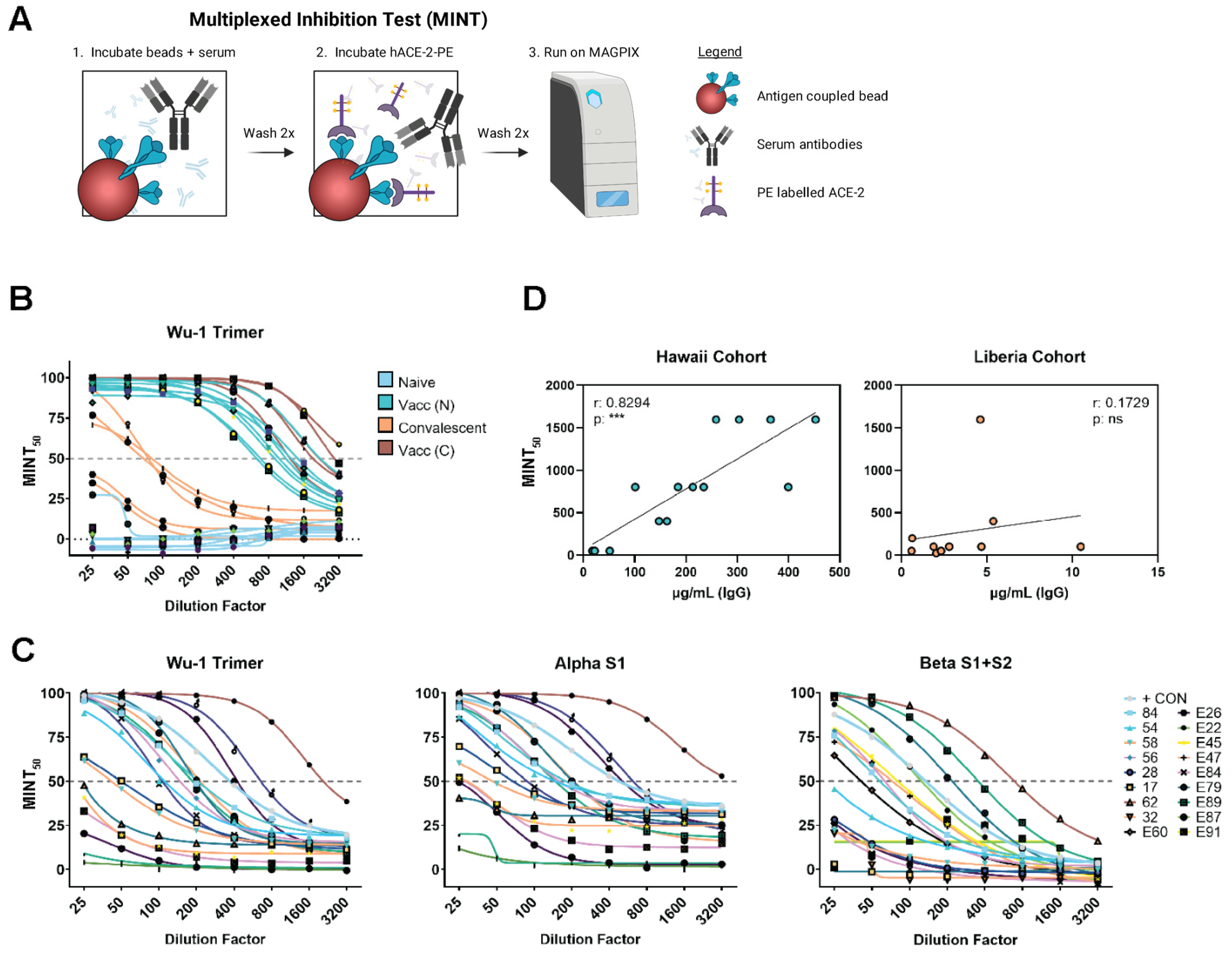
2.5. Multiplexed Inhibition Tests (MINT)
3. Results
3.1. MIA Determines SARS-CoV-2 Serological Status
3.2. Liberian Sera Display Broad Antiviral IgG Reactivity
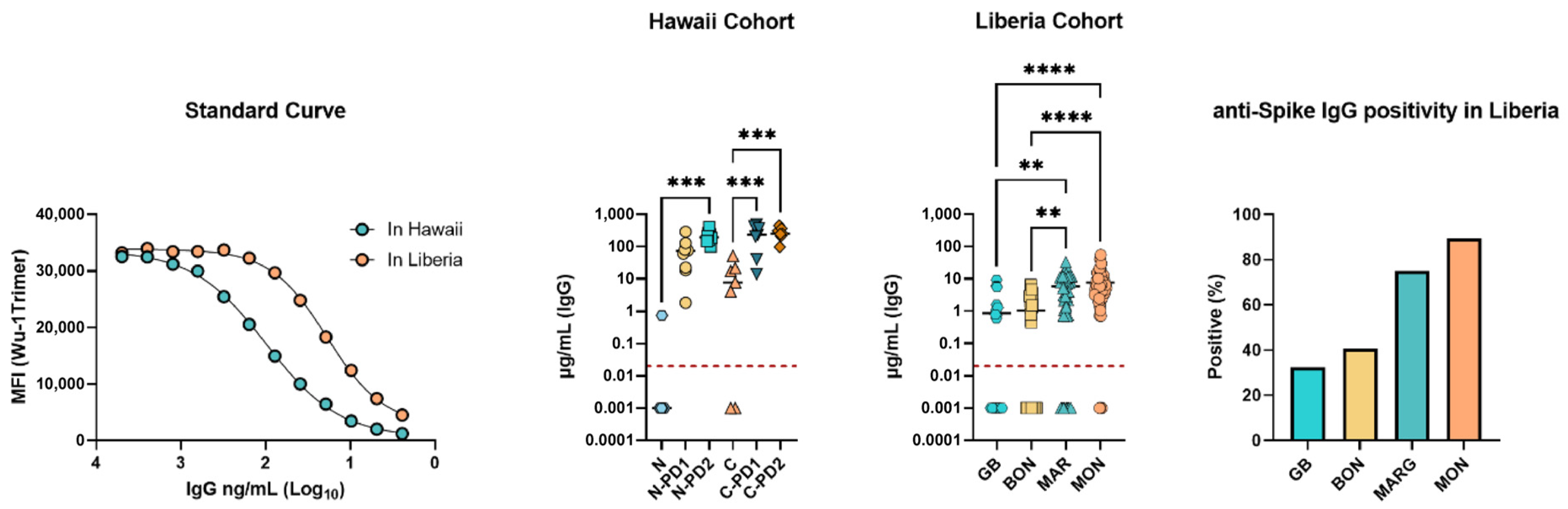
3.3. SARS-CoV-2 Spike (Wuhan Hu-1 Strain) Specific IgG Concentrations
3.4. Multiplexed Inhibition Test (MINT) Reveals Antibody Functionality
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nkengasong, J. Let Africa into the market for COVID-19 diagnostics. Nature 2020, 580, 565–566. [Google Scholar] [CrossRef]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Mertens, P.; De Vos, N.; Martiny, D.; Jassoy, C.; Mirazimi, A.; Cuypers, L.; Van den Wijngaert, S.; Monteil, V.; Melin, P.; Stoffels, K.; et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front. Med. 2020, 7, 225. [Google Scholar] [CrossRef]
- Porte, L.; Legarraga, P.; Vollrath, V.; Aguilera, X.; Munita, J.M.; Araos, R.; Pizarro, G.; Vial, P.; Iruretagoyena, M.; Dittrich, S.; et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020, 99, 328–333. [Google Scholar] [CrossRef]
- Candel González, F.J.; Viñuela-Prieto, J.M.; González Del Castillo, J.; Barreiro García, P.; Fragiel Saavedra, M.; Hernández Píriz, A.; Jiménez Virumbrales, D.; Canora Lebrato, J.; García De Casasola, G.; Gil Prieto, R.; et al. Utility of lateral flow tests in SARS-CoV-2 infection monitorization. Rev. Española De Quimioter. 2020, 33, 258–266. [Google Scholar] [CrossRef]
- Peto, T.; Team, U.C.-L.F.O. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine 2021, 36, 100924. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, T.; Kolstad, L.; Lindahl, J.F.; Albinsson, B.; Bergqvist, A.; Rönnberg, B.; Lundkvist, Å. Diagnostic Potential of a Luminex-Based Coronavirus Disease 2019 Suspension Immunoassay (COVID-19 SIA) for the Detection of Antibodies against SARS-CoV-2. Viruses 2021, 13, 993. [Google Scholar] [CrossRef] [PubMed]
- Medina, L.O.; To, A.; Lieberman, M.M.; Wong, T.A.S.; Namekar, M.; Nakano, E.; Andersen, H.; Yalley-Ogunro, J.; Greenhouse, J.; Higgs, S.; et al. A Recombinant Subunit Based Zika Virus Vaccine Is Efficacious in Non-human Primates. Front. Immunol. 2018, 9, 2464. [Google Scholar] [CrossRef] [PubMed]
- Haun, B.K.; Kamara, V.; Dweh, A.S.; Garalde-Machida, K.; Forkay, S.S.E.; Takaaze, M.; Namekar, M.; Wong, T.A.S.; Bell-Gam Woto, A.E.R.; Humphreys, P.; et al. Serological evidence of Ebola virus exposure in dogs from affected communities in Liberia: A preliminary report. PLoS Negl. Trop. Dis. 2019, 13, e0007614. [Google Scholar] [CrossRef] [PubMed]
- Haun, B.K.; Lai, C.Y.; Williams, C.A.; Wong, T.A.S.; Lieberman, M.M.; Pessaint, L.; Andersen, H.; Lehrer, A.T. CoVaccine HT Adjuvant Potentiates Robust Immune Responses to Recombinant SARS-CoV-2 Spike S1 Immunization. Front. Immunol. 2020, 11, 599587. [Google Scholar] [CrossRef] [PubMed]
- To, A.; Wong, T.A.S.; Lieberman, M.M.; Thompson, K.; Ball, A.H.; Pessaint, L.; Greenhouse, J.; Daham, N.; Cook, A.; Narvaez, B.; et al. A Recombinant Subunit Vaccine Induces a Potent, Broadly Neutralizing, and Durable Antibody Response in Macaques against the SARS-CoV-2 P.1 (Gamma) Variant. ACS Infect. Dis. 2022, 8, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Roe, K.; Nerurkar, P.V.; Namekar, M.; Orillo, B.; Verma, S.; Nerurkar, V.R. Impaired virus clearance, compromised immune response and increased mortality in type 2 diabetic mice infected with West Nile virus. PLoS ONE 2012, 7, e44682. [Google Scholar] [CrossRef] [PubMed]
- Namekar, M.; Kumar, M.; O’Connell, M.; Nerurkar, V.R. Effect of serum heat-inactivation and dilution on detection of anti-WNV antibodies in mice by West Nile virus E-protein microsphere immunoassay. PLoS ONE 2012, 7, e45851. [Google Scholar] [CrossRef] [PubMed]
- Sancilio, A.E.; D’Aquila, R.T.; McNally, E.M.; Velez, M.P.; Ison, M.G.; Demonbreun, A.R.; McDade, T.W. A surrogate virus neutralization test to quantify antibody-mediated inhibition of SARS-CoV-2 in finger stick dried blood spot samples. Sci. Rep. 2021, 11, 15321. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.C.; Tiu, C.; Hu, Z.; Chen, V.C.-W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Wisnewski, A.V.; Liu, J.; Lucas, C.; Klein, J.; Iwasaki, A.; Cantley, L.; Fazen, L.; Campillo Luna, J.; Slade, M.; Redlich, C.A. Development and utilization of a surrogate SARS-CoV-2 viral neutralization assay to assess mRNA vaccine responses. PLoS ONE 2022, 17, e0262657. [Google Scholar] [CrossRef]
- Embregts, C.W.E.; Verstrepen, B.; Langermans, J.A.M.; Boszormenyi, K.P.; Sikkema, R.S.; de Vries, R.D.; Hoffmann, D.; Wernike, K.; Smit, L.A.M.; Zhao, S.; et al. Evaluation of a multi-species SARS-CoV-2 surrogate virus neutralization test. One Health 2021, 13, 100313. [Google Scholar] [CrossRef]
- Ching, L.L.; Tseng, A.C.; Nakano, E.; Salomon, R.C.; Wang, W.K.; Shikuma, C.; Nerurkar, V.R. COVID-19 vaccination and booster induced authentic-virus neutralizing antibody response is superior to SARS-CoV-2 natural infection induced response. J. Clin. Virol. 2022, 152, 105185. [Google Scholar] [CrossRef]
- To, A.; Medina, L.O.; Mfuh, K.O.; Lieberman, M.M.; Wong, T.A.S.; Namekar, M.; Nakano, E.; Lai, C.Y.; Kumar, M.; Nerurkar, V.R.; et al. Recombinant Zika Virus Subunits Are Immunogenic and Efficacious in Mice. mSphere 2018. [Google Scholar] [CrossRef]
- Shobayo, B.; Mishra, M.; Sameroff, S.; Petrosov, A.; Ng, J.; Gokden, A.; Macauley, J.; Jain, K.; Renken, C.; Duworko, J.T.; et al. SARS-CoV-2 Sequence Analysis during COVID-19 Case Surge, Liberia, 2021. Emerg. Infect. Dis. 2021, 27, 3185–3188. [Google Scholar] [CrossRef]
- Barrie, M.B.; Lakoh, S.; Kelly, J.D.; Kanu, J.S.; Squire, J.S.; Koroma, Z.; Bah, S.; Sankoh, O.; Brima, A.; Ansumana, R.; et al. SARS-CoV-2 antibody prevalence in Sierra Leone, March 2021: A cross-sectional, nationally representative, age-stratified serosurvey. BMJ Glob. Health 2021, 6, e007271. [Google Scholar] [CrossRef]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef]
- Akpan, G.E.; Bawo, L.; Amo-Addae, M.; Kennedy, J.; Wesseh, C.S.; Whesseh, F.; Adewuyi, P.; Sanvee-Blebo, L.; Babalola, J.; Sesay, H.W.W.; et al. COVID-19 reinfection in Liberia: Implication for improving disease surveillance. PLoS ONE 2022, 17, e0265768. [Google Scholar] [CrossRef]
- Tang, J.W.; Lam, T.T.; Zaraket, H.; Lipkin, W.I.; Drews, S.J.; Hatchette, T.F.; Heraud, J.M.; Koopmans, M.P.; INSPIRE investigators. Global epidemiology of non-influenza RNA respiratory viruses: Data gaps and a growing need for surveillance. Lancet Infect. Dis. 2017, 17, e320–e326. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Z.; Wang, X.; Zhang, X.; Wang, L.; Lu, Z.; Jia, Z. Association between human coronaviruses’ epidemic and environmental factors on a global scale. Environ. Sci. Pollut. Res. 2022, 29, 14333–14347. [Google Scholar] [CrossRef]
- Edridge, A.W.D.; Kaczorowska, J.; Hoste, A.C.R.; Bakker, M.; Klein, M.; Loens, K.; Jebbink, M.F.; Matser, A.; Kinsella, C.M.; Rueda, P.; et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020, 26, 1691–1693. [Google Scholar] [CrossRef]
- Waterlow, N.R.; van Leeuwen, E.; Davies, N.G.; Group, C.C.-W.; Flasche, S.; Eggo, R.M. How immunity from and interaction with seasonal coronaviruses can shape SARS-CoV-2 epidemiology. Proc. Natl. Acad. Sci. USA 2021, 118, e2108395118. [Google Scholar] [CrossRef]
- Galipeau, Y.; Siragam, V.; Laroche, G.; Marion, E.; Greig, M.; McGuinty, M.; Booth, R.A.; Durocher, Y.; Cuperlovic-Culf, M.; Bennett, S.A.L.; et al. Relative Ratios of Human Seasonal Coronavirus Antibodies Predict the Efficiency of Cross-Neutralization of SARS-CoV-2 Spike Binding to ACE2. EBioMedicine 2021, 74, 103700. [Google Scholar] [CrossRef] [PubMed]
- Sagar, M.; Reifler, K.; Rossi, M.; Miller, N.S.; Sinha, P.; White, L.F.; Mizgerd, J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Investig. 2021. [Google Scholar] [CrossRef] [PubMed]
- Borrega, R.; Nelson, D.K.S.; Koval, A.P.; Bond, N.G.; Heinrich, M.L.; Rowland, M.M.; Lathigra, R.; Bush, D.J.; Aimukanova, I.; Phinney, W.N.; et al. Cross-Reactive Antibodies to SARS-CoV-2 and MERS-CoV in Pre-COVID-19 Blood Samples from Sierra Leoneans. Viruses 2021, 13, 2325. [Google Scholar] [CrossRef] [PubMed]
- Aoussi, E.B.; Ehui, E.; Kassi, N.A.; Kouakou, G.; Nouhou, Y.; Adjogoua, E.V.; Eholie, S.; Bissagnene, E. Seven native cases of dengue in Abidjan, Ivory Coast. Med. Mal. Infect. 2014, 44, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Dariano, D.F.; Taitt, C.R.; Jacobsen, K.H.; Bangura, U.; Bockarie, A.S.; Bockarie, M.J.; Lahai, J.; Lamin, J.M.; Leski, T.A.; Yasuda, C.; et al. Surveillance of Vector-Borne Infections (Chikungunya, Dengue, and Malaria) in Bo, Sierra Leone, 2012–2013. Am. J. Trop. Med. Hyg. 2017, 97, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- De Araújo Lobo, J.M.; Mores, C.N.; Bausch, D.G.; Christofferson, R.C. Short Report: Serological Evidence of Under-Reported Dengue Circulation in Sierra Leone. PLoS Neglected Trop. Dis. 2016, 10, e0004613. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Were, F. The dengue situation in Africa. Paediatr. Int. Child. Health 2012, 32 (Suppl. S1), 18–21. [Google Scholar] [CrossRef]
- Russo, G.; Subissi, L.; Rezza, G. Chikungunya fever in Africa: A systematic review. Pathog. Glob. Health 2020, 114, 136–144. [Google Scholar] [CrossRef]
- Conzelmann, C.; Gilg, A.; Gross, R.; Schutz, D.; Preising, N.; Standker, L.; Jahrsdorfer, B.; Schrezenmeier, H.; Sparrer, K.M.J.; Stamminger, T.; et al. An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antivir. Res. 2020, 181, 104882. [Google Scholar] [CrossRef]
- Muller, K.; Girl, P.; von Buttlar, H.; Dobler, G.; Wolfel, R. Comparison of two commercial surrogate ELISAs to detect a neutralising antibody response to SARS-CoV-2. J. Virol. Methods 2021, 292, 114122. [Google Scholar] [CrossRef]
- Wouters, E.; Verbrugghe, C.; Devloo, R.; Debruyne, I.; De Clippel, D.; Van Heddegem, L.; Van Asch, K.; Van Gaver, V.; Vanbrabant, M.; Muylaert, A.; et al. A novel competition ELISA for the rapid quantification of SARS-CoV-2 neutralizing antibodies in convalescent plasma. Transfusion 2021, 61, 2981–2990. [Google Scholar] [CrossRef]
- Grunau, B.; Prusinkiewicz, M.; Asamoah-Boaheng, M.; Golding, L.; Lavoie, P.M.; Petric, M.; Levett, P.N.; Haig, S.; Barakauskas, V.; Karim, M.E.; et al. Correlation of SARS-CoV-2 Viral Neutralizing Antibody Titers with Anti-Spike Antibodies and ACE-2 Inhibition among Vaccinated Individuals. Microbiol. Spectr. 2022, 10, e0131522. [Google Scholar] [CrossRef]
- Kohmer, N.; Westhaus, S.; Ruhl, C.; Ciesek, S.; Rabenau, H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020, 129, 104480. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Ko, J.-H.; Park, J.; Moon, H.-W.; Baek, J.Y.; Jung, S.; Lim, H.-Y.; Kim, K.-C.; Huh, K.; Cho, S.Y.; et al. Estimating the Neutralizing Effect and Titer Correlation of Semi-Quantitative Anti-SARS-CoV-2 Antibody Immunoassays. Front. Cell. Infect. Microbiol. 2022, 12, 822599. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Jung, J.; Lee, J.H.; Lee, D.-G.; Kim, Y.B.; Oh, E.-J. Comparison of Six Serological Immunoassays for the Detection of SARS-CoV-2 Neutralizing Antibody Levels in the Vaccinated Population. Viruses 2022, 14, 946. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Hall, T.; Ssewanyana, I.; Oulton, T.; Patterson, C.; Vasileva, H.; Singh, S.; Affara, M.; Mwesigwa, J.; Correa, S.; et al. Optimisation and standardisation of a multiplex immunoassay of diverse Plasmodium falciparum antigens to assess changes in malaria transmission using sero-epidemiology. Wellcome Open Res. 2019, 4, 26. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haun, B.K.; To, A.; Williams, C.A.; Ball, A.; Fong, K.; Wong, T.A.S.; Shobayo, B.; Teahton, J.; Ching, L.; Kamara, V.; et al. A Serological Multiplexed Immunoassay (MIA) Detects Antibody Reactivity to SARS-CoV-2 and Other Viral Pathogens in Liberia and Is Configurable as a Multiplexed Inhibition Test (MINT). Immuno 2024, 4, 108-124. https://doi.org/10.3390/immuno4010007
Haun BK, To A, Williams CA, Ball A, Fong K, Wong TAS, Shobayo B, Teahton J, Ching L, Kamara V, et al. A Serological Multiplexed Immunoassay (MIA) Detects Antibody Reactivity to SARS-CoV-2 and Other Viral Pathogens in Liberia and Is Configurable as a Multiplexed Inhibition Test (MINT). Immuno. 2024; 4(1):108-124. https://doi.org/10.3390/immuno4010007
Chicago/Turabian StyleHaun, Brien K., Albert To, Caitlin A. Williams, Aquena Ball, Karalyn Fong, Teri Ann S. Wong, Bode Shobayo, Julius Teahton, Lauren Ching, Varney Kamara, and et al. 2024. "A Serological Multiplexed Immunoassay (MIA) Detects Antibody Reactivity to SARS-CoV-2 and Other Viral Pathogens in Liberia and Is Configurable as a Multiplexed Inhibition Test (MINT)" Immuno 4, no. 1: 108-124. https://doi.org/10.3390/immuno4010007
APA StyleHaun, B. K., To, A., Williams, C. A., Ball, A., Fong, K., Wong, T. A. S., Shobayo, B., Teahton, J., Ching, L., Kamara, V., Tekah, D. M., Humphrey, P., Berestecky, J., Nerurkar, V. R., & Lehrer, A. T. (2024). A Serological Multiplexed Immunoassay (MIA) Detects Antibody Reactivity to SARS-CoV-2 and Other Viral Pathogens in Liberia and Is Configurable as a Multiplexed Inhibition Test (MINT). Immuno, 4(1), 108-124. https://doi.org/10.3390/immuno4010007






