Abstract
COVID-19 has presented itself as a challenging task to medical teams and researchers throughout the world, since the outbreak of SARS-CoV-2 started in the Chinese city of Wuhan. To this day, there are still new variants emerging, and the knowledge about the mechanisms used by the virus to infect cells and perpetuate itself are still not well understood. The scientific community is still trying to catch up with the velocity of new variants and, consequently, the new physiological pathways that appear along with it. It is known that the new coronavirus plays a role in changing many molecular pathways to take control of the infected cells. Many of these pathways are related to control genomic expression of certain genes by epigenetic ways, allowing the virus to modulate immune responses and cytokines production. The let-7 family of microRNAs, for instance, are known to promote increased viral fusion in the target cell through a mechanism involving the transmembrane serine protease 2 (TMPRSS2). It was also demonstrated they are able to increase the inflammatory activity through the NF-κB/IL-6/let-7/LIN-28 axis. In addition, let-7 overexpression led to a reduction in inflammatory cytokines and chemokines expression (IL-6, IL-8 and TNF-α). Interestingly, the cytokines modulated by the let-7 family are related to COVID-19-induced cytokine storm observed in patients undergoing clinical phase three. Thus, let-7 can be considered a novel and attractive biomarker for therapeutic purpose. Based on that, the present study aims to critically analyze the immunopathological mechanisms of the microRNA let-7 in the infection caused by SARS-CoV-2.
1. Introduction
In December 2019, the previously unknown Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus spread among the population of Wuhan, China. The rapid global advance of the virus and the thousands of deaths caused by coronavirus disease (COVID-19) have prompted the World Health Organization (WHO) to declare a state of pandemic on 12 March 2020 [1,2,3].
With a re-emerging pathogen such as the SARS-CoV-2, it remains absolutely necessary to gain a better understanding of the precise mechanisms of the SARS-CoV-2 pathophysiology and future viral variants [4]. In this context, the host immune response to the virus appears to play a key role in the disease pathogenesis and clinical presentation. An excessive inflammatory reaction, characterized by a marked pro-inflammatory cytokine release, is remarkable in patients with severe COVID-19. Thus, leading to immune abnormalities which may lead to persisted infections and septic shock [5,6,7].
Coronavirus has a 5′cap structure and 3′polyA tail. The spike glycoprotein (S), envelope (E), membrane (M) and nucleocapsid (N) are structural proteins in coronaviruses. S protein is cleaved to S1 and S2 subunits. S2 facilitates the entry into target cells [8,9,10]. A major host protease, named transmembrane serine protease 2 (TMPRSS2), enables SARS-CoV-2 entry into host cells by priming the spike protein. SARS-CoV-2 uses the angiotensin converting enzyme 2 (ACE2) as a receptor for host cell attachment; once attached, the viral spike protein is cleaved by TMPRSS2 to allow fusion of the viral and cellular membranes [11,12]. The host protease TMPRSS2 plays a critical role in facilitating SARS-CoV-2 entry into host cells by priming the viral spike protein. Once the spike protein attaches to the host receptor ACE2, TMPRSS2 cleaves it, allowing fusion of the viral and cellular membranes. This process enables viral entry and subsequent replication, leading to the development of COVID-19. However, in some individuals, a dysregulated immune response occurs, resulting in a cytokine storm. This immune response involves the release of large amounts of pro-inflammatory cytokines and chemokines by immune effector cells, which can cause severe damage to host tissues and organs. The dysregulated immune response and cytokine storm are major causes of death in COVID-19. Therefore, TMPRSS2 may play a role not only in viral entry but also in the development of cytokine storm [13].
In a cytokine storm, the immune system produces an excessive amount of pro-inflammatory cytokines, such as interleukin-6 (IL-6), interleukin-1 beta (IL-1β), and tumor necrosis factor-alpha (TNF-α), among others. These cytokines can cause widespread inflammation throughout the body, which can lead to tissue damage and organ failure. In COVID-19, the cytokine storm is thought to contribute to the development of acute respiratory distress syndrome (ARDS), which is a severe lung condition that can lead to respiratory failure [14]. IL-6 is also involved in systemic inflammation during infection, which is probably influenced by pre-existing comorbidities [15]. This is followed by the infiltration of macrophages and neutrophils into the lung tissue, which results in a cytokine storm [16,17,18]. According to Jiang et al. (2022), further studies are still needed to fully elucidate the mechanism behind the cytokine storm induced by SARS-CoV-2 infection and thus provide new targets for therapeutic interventions [19].
The NF-κB signal transduction pathway is a common pathway centrally involved in the generation of pro-inflammatory cytokine and chemokine cascades in COVID-19 [20,21,22]. Inhibition of the NF-κB pathway increased survival rates in mice infected with SARS-CoV, due the fact that NF-κB signaling pathway activation is one of the major contributions to the inflammation induced upon SARS-CoV infection [23]. On a more molecular level, increased NF-κB activity leads to increased expression of Lin28, a gene that encodes an RNA-binding protein that plays a key role in regulating gene expression and developmental processes in many organisms, including humans. Lin28 has been shown to inhibit the maturation of let-7 family miRNAs, miRNAs that normally inhibits NF-κB transcripts and activators such as IL-6 [24].
MiRNAs can inhibit the viral translation after the attachment of miRNAs to 3′-UTR of the viral genome without affecting the expression of human genes [25,26]. In addition, the let-7 family is among the first microRNAs discovered. Most of the let-7 regulatory proteins are well studied in development, proliferation, differentiation, and cancer, but their role in inflammation and in infectious diseases such as COVID-19 is not well understood [27,28,29]. The present study aimed to systematically review the studies that were able to describe the role of let-7 in COVID-19, and to provide a basis for future research aimed at therapeutics in this area.
2. Materials and Methods
An extensive search was conducted in the MedLine database to identify studies addressing the relationship between let-7 family miRNAs and the pathophysiology of COVID-19. A keyword search was performed as follows: “COVID-19 or SARS-CoV-2” AND “let-7 or let7”. The database was searched on 25 April 2022, for entries from 1 January 2020 to 24 April 2022. Publication format was limited to peer-reviewed journal articles (as a filter for quality resulting from the peer review process), including all types of publications. We included publications from any country in published in English language.
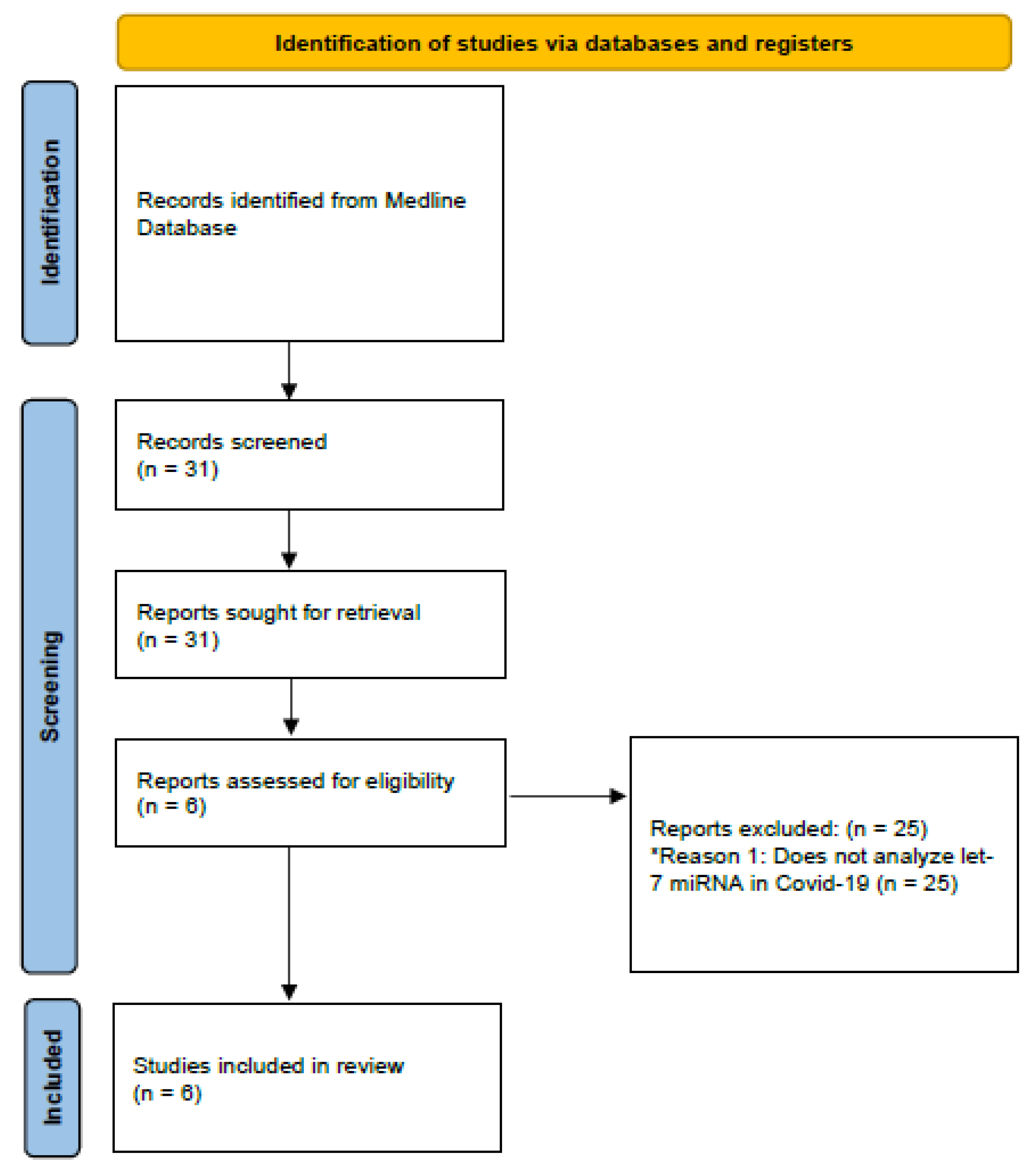
Subsequently, exclusion and inclusion criteria were applied. Studies that addressed the association between let-7 and the pathophysiology of COVID-19 were included, as well as studies performed in adults (older than 18 years). Studies that did not match the criteria described above were excluded (Figure 1).
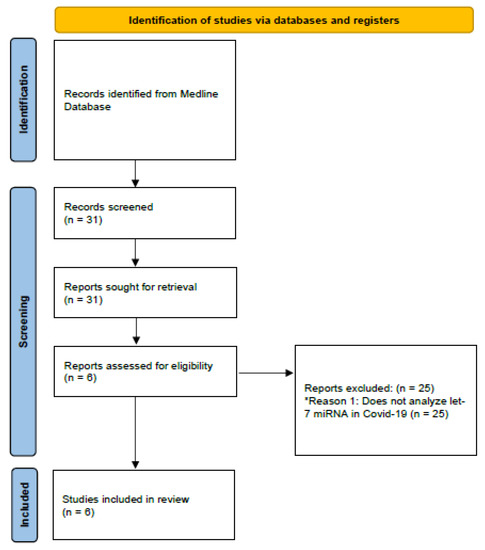
Figure 1.
Diagram demonstrating the methodology used in the present review. * Reason 1: Reason for excluding articles.
Risk of Bias
The Risk of Bias in Non-randomized Studies–of Intervention (ROBINS-I) Tool was used to assess the methodological quality of all studies performed in human beings by evaluating the extent to which they addressed the possibility of bias in seven areas of study design [30]. Two researchers (RLP, GRA) independently assessed each included study and any uncertainty regarding the quality of publications was resolved through discussion among them.
3. Results
The search yielded 31 articles. After applying the search filter and considering the inclusion criteria defined for the present study, six studies capable of identifying the cellular mechanisms of let-7 in COVID-19 were selected. Table 1 describes the studies included in the review. Table 2 presents a comparative summary of all the experiments included in the study.

Table 1.
Articles included in the review results.

Table 2.
Characteristics and main findings of the analyzed articles.
3.1. TMPRSS2
Studies have predicted that let-7 family miRNAs target the 3’ UTR of the TMPRSS2 gene, a membrane protein that plays a crucial role during viral entry into the host cell, thus inhibiting its expression. Furthermore, it was reported that blocking the transcription of let-7 family members such as hsa-let-7e and hsa-let-7a-5p led to increased expression of TMPRSS2 gene [31,35].
3.2. NF-κB and TLR4
An in silico analysis of the TLR4 and NF-κB signaling pathways was performed and pointed out that miR-let-7b was among the top-ranked regulators of these inflammatory pathways, targeting 15 and 14 genes within the TLR4 and NF-κB pathways, respectively. Subsequently, animal models were used to investigate the role of miR-let-7b in inflammation in vivo. Animal models were induced to sepsis by cecal ligation and puncture. In sepsis-induced rat models, compared to control (sham-operation) the expression of nuclear TLR4 and NF-κB increased, while IκBα, a protein that participates in NF-κB inhibition, decreased. While in sepsis-induced mouse models group that received miR-let-7b-agomir treatment (10 nmol miR-let-7b-agomir per mouse suspended in 200 µL saline) was found to significantly decrease nuclear TLR4 and NF-κB and increased IκBα protein levels in neutrophils compared to those that did not receive the treatment. Furthermore, the group treated with miR-let-7b agomir significantly decreased the levels of IL-6 in mouse serum compared to the CLP group. Overall, these results demonstrated that miR-let-7b inhibited CLP-induced inflammation partially through the miR-let-7b/TLR4/NF-κB axis in neutrophils [33].
3.3. IL-6
A case–control experiment performed using 37 COVID-19 patients recruited from four local hospitals in Guangdong revealed increased IL-6 and decreased let-7 family miRNAs in patients of both the severe and mild groups compared to healthy subjects. The mild and severe groups also showed increased expression of the transcription factor NF-κB [34].
In this context, cocultivation of miR-let-7b mimetics with human neutrophils preconditioned by LPS was performed. Neutrophils can be activated by LPS to a pro-inflammatory state. Results with RT-qPCR and ELISA showed that after pre-treatment with LPS and incubation with miR-let-7b, neutrophils produced dramatically fewer pro-inflammatory cytokines, including IL-6, IL-8, and TNFα, and more anti-inflammatory cytokine IL-10 [33].
Additionally, studies have shown that TH1 cells play a crucial role in the immune response to COVID-19. TH1 cells are responsible for producing pro-inflammatory cytokines such as interferon-gamma (IFN-γ), which can help to clear the virus and prevent its spread. However, in severe cases of COVID-19, the immune response can become dysregulated, leading to a cytokine storm and potentially fatal complications. Let-7 miRNAs have been shown to regulate the expression of several pro-inflammatory cytokines, including IFN-γ, interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-α). Overexpression of pri-let-7a and pri-let-7c in THP1 cells has been observed to reduce the mRNA levels of IL-6 and other cytokines and chemokines associated with SARS-CoV-2, such as IL-1β, IL-8, CCL2, GM-CSF, and TNFα. Conversely, a reduction in let-7 levels has been shown to increase the levels of these pro-inflammatory cytokines, suggesting that let-7 may play a crucial role in regulating the immune response to COVID-19 [32,37].
3.4. ACE2
An experiment conducted with 31 obese women in New Zealand showed a negative correlation between miR-let-7b and ACE2. Individuals with higher levels of expressed mir-let-7b exhibited lower levels of expressed ACE2 [36].
3.5. Risk of Bias and Quality Assessment
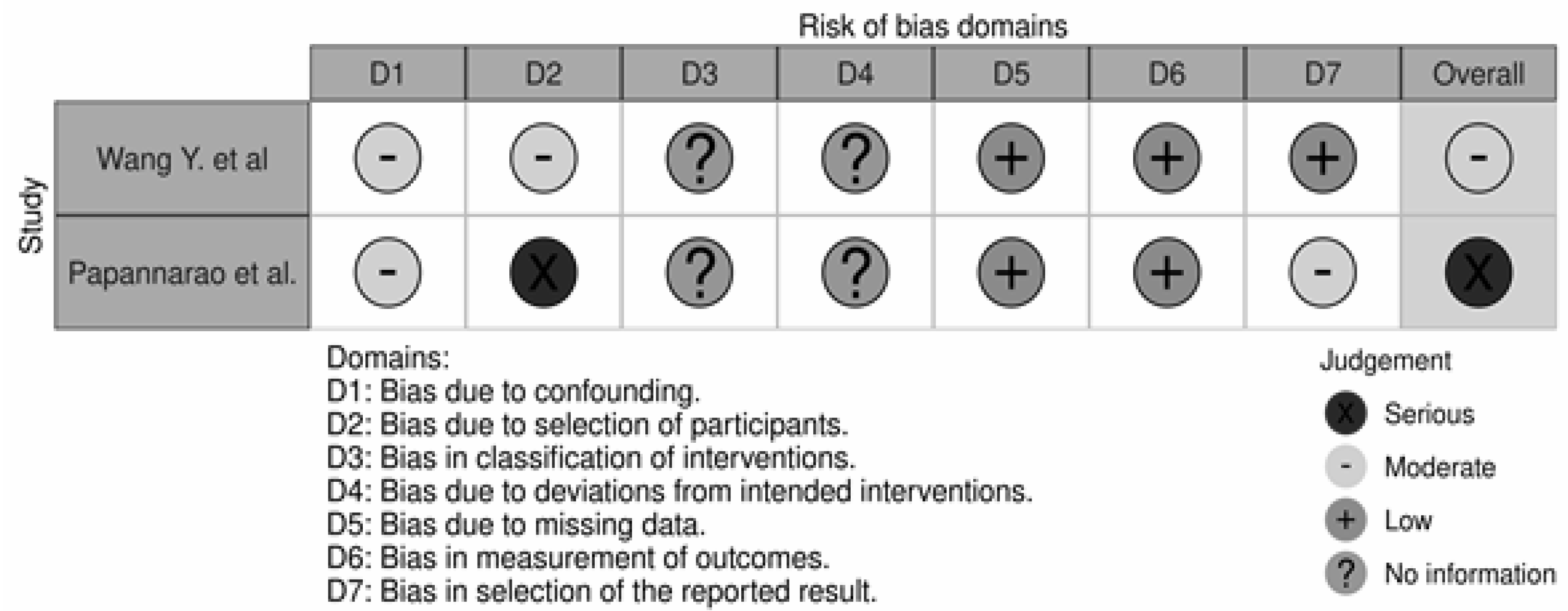
The overall quality of the studies was poor (Figure 2). The study by Wang Y. et al. had a moderate risk of bias regarding participant selection and confounding. The participant selection by Papannarao et al. had a serious risk of bias; the major factor for this assessment concerned the fact that the authors selected only female participants in the study design. Thus, of the two studies performed in humans included in our analysis, one was judged to have a moderate overall risk of bias, and one was judged to have a serious risk of bias [34,36].
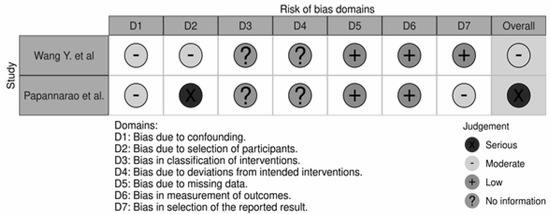
Figure 2.
Risk of Bias assessment of the two clinical trials included in our analysis [34,36].
4. Discussion
MiRNA is a type of direct and potent regulator of gene expression. MiRNA controls gene expression by binding any regions suitable for interaction that can be located in DNA and RNA. The interplay between miRNA and other biomolecules is responsible for the homeostasis of a living organism [38]. Many aspects of the miRNA let-7 intersect with the immunopathology of COVID-19. The present review identified six experiments testing hypotheses about the role of let-7 in COVID-19 pathology, and these were critically summarized in the present article. Only two of these have been performed in humans. The Wang Y. et al. trial has the advantage of having dichotomous groups and recruiting patients from four different hospitals [34]. However, the low number of patients may influence the accuracy of the results.
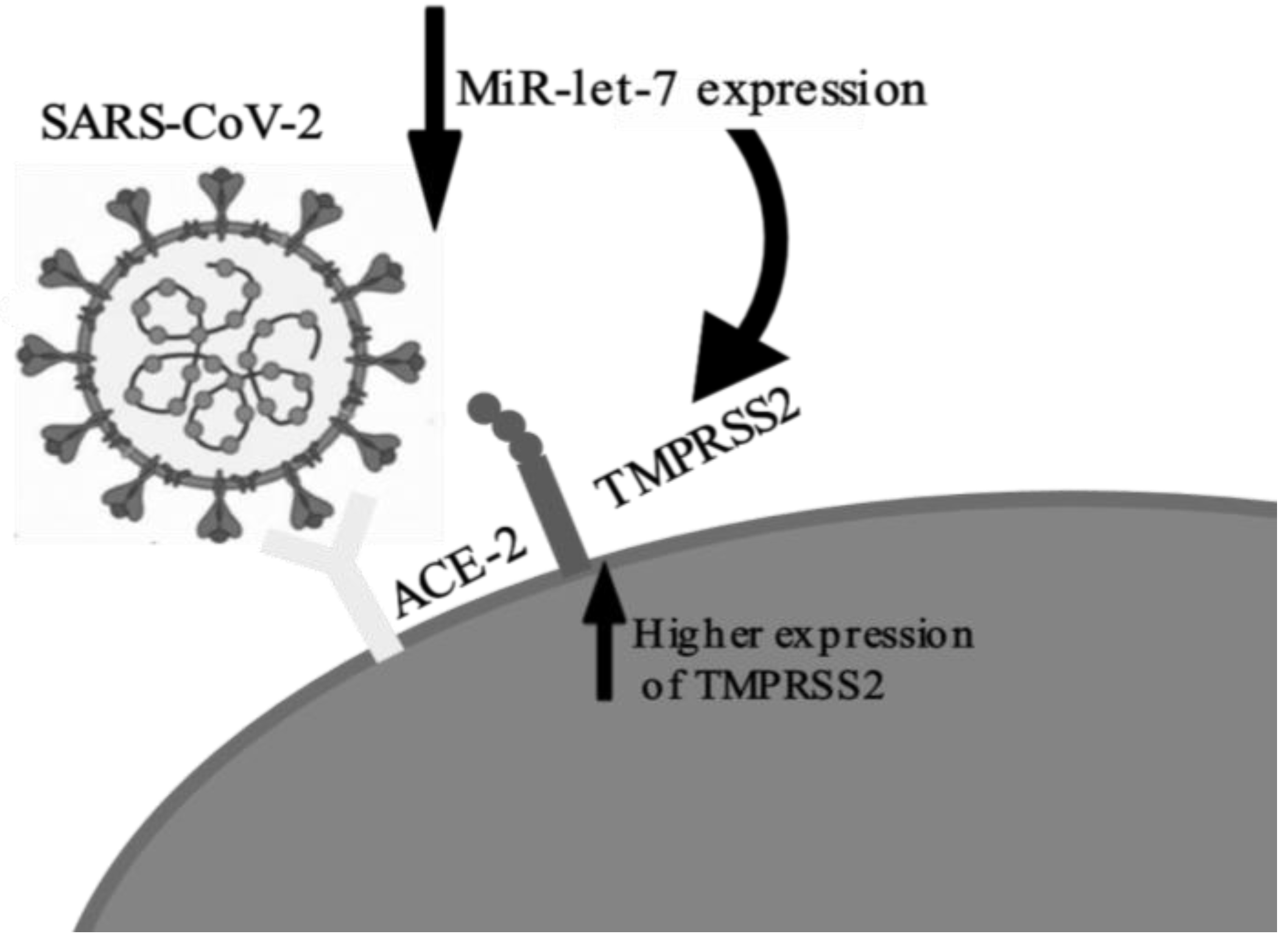
Regarding the findings of each study, the most frequent finding was the increased expression of pro-inflammatory cytokines, especially IL-6, correlated with the suppression of let-7, generating the mechanism for the cytokine storm [33,34,35]. These findings are consistent with the literature of miRNA let-7 [38]. Most of the studies assessed were laboratory-based. They showed plausible correlations between aspects of COVID-19 immunopathology with let-7. Increased expression of TMPRSS2 via repression of let-7 (Figure 3) was cited by two authors [31,35]; this implies an increase in viral uptake into the target cells via two mechanisms: by the cleavage of SARS-S, which activates the S protein for membrane fusion, and by the cleavage of ACE2 [39,40]. The internalization and replication of virus subsequently causes degradation of membrane-bound ACE2 receptors, which in turn causes increase in angiotensin II and the angiotensin type 1 receptor, resulting in an inflammatory immune response [41]. Thus, decreased let-7 expression may play a role in increasing the viral load of the disease by facilitating virus entry into the host cells through increased expression of TMPRSS2.
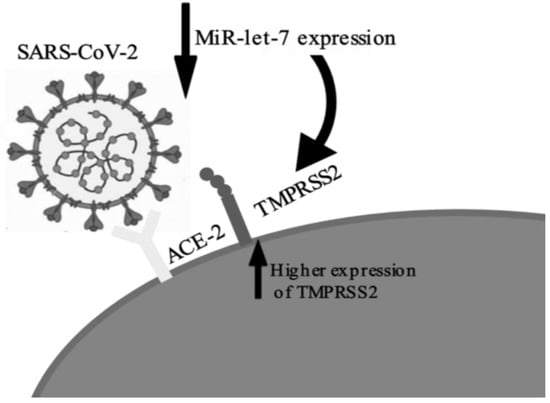
Figure 3.
Representation of let-7 suppression by COVID-19 and the subsequent increase in TMPRSS2 expression.
The let-7 suppression also leads to increased activation of the TLR4/NF-κB pathway [33,42,43,44]. Activated NF-κB transcription factors are a trigger for the expression of a wide variety of cytokines, (e.g., IL-1, IL-2, IL-6 and TNF-α) that can induce an inflammatory programming in resident macrophages and recruit activated monocytes and T cells to the lungs [45,46]. In addition, one of the most highly induced NF-κB-dependent cytokines is IL-6; in patients with COVID-19, elevated IL-6 levels are associated with worse clinical outcomes [47]. Activation of NF-κB promotes the synthesis of the microRNA binding protein LIN-28, which reduces the synthesis of mature let-7, thus constituting an inflammatory loop involving NF-κB/IL-6/let-7/ACE2 [48,49]. These findings are plausible to explain the hyperinflammatory state and cytokine storm induced in severe cases of the disease, which results in tissue damage, acute respiratory distress (ARDS) and multiple organ failure [50,51].
Furthermore, overexpression of let-7 is able to suppress the hyperinflammatory state. Discovered by Xie C., C1632 is a small molecule that serves as a let-7 stimulator, capable of positively regulating let-7 and therefore reducing viral replication and the secretion of pro-inflammatory cytokines. This molecule had attractive results in in vitro studies as a potential therapeutic for COVID-19 [32]. Like C1632, other therapeutic targets may have therapeutic potential through the regulation of let-7, and this is an attractive biomarker for future therapeutic research.
The immunopathological mechanisms of COVID-19 are complex and pose a challenge to researchers. This study provides a critical analysis of let-7 miRNA in the context of COVID-19, serving as a basis for further research in the area. Our paper is the first comprehensive critical analysis published on let-7 and COVID-19. A limitation of our study was the low number of experiments performed, resulting in a low number of articles for analysis, and the low quality of studies performed in humans. As a strong point, our result has good immunopathological plausibility to explain clinical findings of the disease, and it provides reasonable data for future research in this area.
5. Conclusions
Six published experiments relating to let-7 and COVID-19 were identified. Some studies exhibited good accuracy for the correlation of let-7 in the immunopathology of COVID-19. The let-7 miRNA suppression generated in severe COVID-19 implies increased viral uptake by the target cell via increased TMPRSS2 and causes increased inflammation through the NF-κB/IL-6/let-7/LIN-28 axis. The future of let-7 targeted therapy is attractive, but there is a long way to go to obtain a clinically safe drug. Thus, future researchers may successfully set a direction in primary research by targeting let-7 as a therapeutic target.
Author Contributions
R.B.d.O., developed the idea, conceptualization, validation, writing (review and editing), supervision and project administration; R.L.P., performed the conceptualization, methodology, formal analysis, investigation, data curation and writing (original draft); G.d.R.A., performed methodology, formal analysis, investigation and data curation. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Luiz Fernando Kehl for critically contribute to the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.-C.; Wang, C.-B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef]
- Esakandari, H.; Nabi-Afjadi, M.; Fakkari-Afjadi, J.; Farahmandian, N.; Miresmaeili, S.-M.; Bahreini, E. A comprehensive review of COVID-19 characteristics. Biol. Proced. Online 2020, 22, 19. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Pandit, P.; McArthur, A.G.; Banerjee, A.; Mossman, K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021, 18, 166. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Pozzoli, U.; Mozzi, A.; Arrigoni, F.; De Gioia, L.; Clerici, M.; Sironi, M. Dating the Emergence of Human Endemic Coronaviruses. Viruses 2022, 14, 1095. [Google Scholar] [CrossRef]
- Fani, M.; Zandi, M.; Ebrahimi, S.; Soltani, S.; Abbasi, S. The role of miRNAs in COVID-19 disease. Future Virol. 2021, 16, 301–306. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Nikolaidis, M.; Tryfonopoulou, E.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G. The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2. Viruses 2022, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; El Naggar, R.F.; Clayton, E.; Munir, M. Structural and functional insights into non-structural proteins of coronaviruses. Microb. Pathog. 2020, 150, 104641. [Google Scholar] [CrossRef] [PubMed]
- Schönfelder, K.; Breuckmann, K.; Elsner, C.; Dittmer, U.; Fistera, D.; Herbstriet, F.; Risse, J.; Schmidt, K.; Sutharsan, S.; Taube, C.; et al. Transmembrane serine protease 2 Polymorphisms and Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Type 2 Infection: A German Case-Control Study. Front. Genet. 2021, 12, 585. [Google Scholar] [CrossRef] [PubMed]
- Akbasheva, O.; Spirina, L.; Dyakov, D.; Masunova, N. Proteoliz i defitsit α1-proteinaznogo ingibitora pri infektsii SARS-CoV-2 [Proteolysis and deficiency of α1-proteinase inhibitor in SARS-CoV-2 infection]. Biomeditsinskaya Khimiya 2022, 68, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Tahaghoghi-Hajghorbani, S.; Zafari, P.; Masoumi, E.; Rajabinejad, M.; Jafari-Shakib, R.; Hasani, B.; Rafiei, A. The role of dysregulated immune responses in COVID-19 pathogenesis. Virus Res. 2020, 290, 198197. [Google Scholar] [CrossRef]
- Luo, X.; Zhu, Y.; Mao, J.; Du, R. T cell immunobiology and cytokine storm of COVID-19. Scand. J. Immunol. 2021, 93, e12989. [Google Scholar] [CrossRef]
- Jones, S.A.; Hunter, C.A. Is IL-6 a key cytokine target for therapy in COVID-19? Nat. Rev. Immunol. 2021, 21, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Potere, N.; Batticciotto, A.; Vecchié, A.; Porreca, E.; Cappelli, A.; Abbate, A.; Dentali, F.; Bonaventura, A. The role of IL-6 and IL-6 blockade in COVID-19. Expert Rev. Clin. Immunol. 2021, 17, 601–618. [Google Scholar] [CrossRef]
- Gubernatorova, E.O.; Gorshkova, E.A.; Polinova, A.I.; Drutskaya, M.S. IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020, 53, 13–24. [Google Scholar] [CrossRef]
- Jiang, Y.; Rubin, L.; Peng, T.; Liu, L.; Xing, X.; Lazarovici, P.; Zheng, W. Cytokine storm in COVID-19: From viral infection to immune responses, diagnosis and therapy. Int. J. Biol. Sci. 2022, 18, 459–472. [Google Scholar] [CrossRef]
- Kircheis, R.; Haasbach, E.; Lueftenegger, D.; Heyken, W.T.; Ocker, M.; Planz, O. NF-κB Pathway as a Potential Target for Treatment of Critical Stage COVID-19 Patients. Front. Immunol. 2020, 11, 3446. [Google Scholar] [CrossRef]
- Aydemir, M.N.; Aydemir, H.B.; Korkmaz, E.M.; Budak, M.; Cekin, N.; Pinarbasi, E. Computationally predicted SARS-CoV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021, 22, 101012. [Google Scholar] [CrossRef] [PubMed]
- Attiq, A.; Yao, L.J.; Afzal, S.; Khan, M.A. The triumvirate of NF-κB, inflammation and cytokine storm in COVID-19. Int. Immunopharmacol. 2021, 101, 108255. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Nieto-Torres, J.L.; Regla-Nava, J.A.; Jimenez-Guardeño, J.M.; Fernandez-Delgado, R.; Castaño-Rodriguez, C.; Perlman, S.; Enjuanes, L. Inhibition of NF-κB-Mediated Inflammation in Severe Acute Respiratory Syndrome Coronavirus-Infected Mice Increases Survival. J. Virol. 2014, 88, 913–924. [Google Scholar] [CrossRef]
- Mills, W.T., IV; Nassar, N.N.; Ravindra, D.; Li, X.; Meffert, M.K. Multi-Level Regulatory Interactions between NF-κB and the Pluripotency Factor Lin28. Cells 2020, 9, 2710. [Google Scholar] [CrossRef] [PubMed]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLOS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Han, H.-S. Coronavirus disease 2019 and mRNA vaccines: What’s next–miRNA? Clin. Exp. Pediatr. 2022, 65, 302–303. [Google Scholar] [CrossRef]
- Bernstein, D.L.; Jiang, X.; Rom, S. let-7 microRNAs: Their Role in Cerebral and Cardiovascular Diseases, Inflammation, Cancer, and Their Regulation. Biomedicines 2021, 9, 606. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. In Methods in Molecular Biology; Springer: Berlin, Germany, 2017; Volume 1509, pp. 1–10. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, S.; Shkurnikov, M.; Turchinovich, A.; Knyazev, E.; Tonevitsky, A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS ONE 2020, 15, e0235987. [Google Scholar] [CrossRef]
- Xie, C.; Chen, Y.; Luo, D.; Zhuang, Z.; Jin, H.; Zhou, H.; Li, X.; Lin, H.; Zheng, X.; Zhang, J.; et al. Therapeutic potential of C1632 by inhibition of SARS-CoV-2 replication and viral-induced inflammation through upregulating let-7. Signal Transduct. Target. Ther. 2021, 6, 84. [Google Scholar] [CrossRef]
- Chen, B.; Han, J.; Chen, S.; Xie, R.; Yang, J.; Zhou, T.; Zhang, Q.; Xia, R. MicroLet-7b Regulates Neutrophil Function and Dampens Neutrophilic Inflammation by Suppressing the Canonical TLR4/NF-κB Pathway. Front. Immunol. 2021, 12, 856. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Zhang, L.; Sun, H.-X.; Zhang, Z.; Xu, J.; Xu, Y.; Lin, Y.; Zhu, A.; Luo, Y.; et al. Plasma cell-free RNA characteristics in COVID-19 patients. Genome Res. 2022, 32, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, D.; Fiselier, A.; Kovalchuk, I.; Kovalchuk, O. New AKT-dependent mechanisms of anti-COVID-19 action of high-CBD Cannabis sativa extracts. Cell Death Discov. 2022, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Papannarao, J.B.; Schwenke, D.O.; Manning, P.; Katare, R. Upregulated miR-200c is associated with downregulation of the functional receptor for severe acute respiratory syndrome coronavirus 2 ACE2 in individuals with obesity. Int. J. Obes. 2021, 46, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhang, S.; Amahong, K.; Sun, X.; Lian, X.; Liu, J.; Sun, H.; Lou, Y.; Zhu, F.; Qiu, Y. The miRNA: A small but powerful RNA for COVID-19. Briefings Bioinform. 2021, 22, 1137–1149. [Google Scholar] [CrossRef]
- Sung, S.-Y.; Liao, C.-H.; Wu, H.-P.; Hsiao, W.-C.; Wu, I.-H.; Jinpu, Y.; Lin, S.-H.; Hsieh, C.-L. Loss of Let-7 MicroRNA Upregulates IL-6 in Bone Marrow-Derived Mesenchymal Stem Cells Triggering a Reactive Stromal Response to Prostate Cancer. PLoS ONE 2013, 8, e71637. [Google Scholar] [CrossRef]
- Zipeto, D.; da Fonseca Palmeira, J.; Argañaraz, G.A.; Argañaraz, E.R. ACE2/ADAM17/TMPRSS2 Interplay May Be the Main Risk Factor for COVID-19. Front. Immunol. 2020, 11, 2642. [Google Scholar] [CrossRef]
- Pollard, C.A.; Morran, M.P.; Nestor-Kalinoski, A.L. The COVID-19 pandemic: A global health crisis. Physiol. Genom. 2020, 52, 549–557. [Google Scholar] [CrossRef]
- Wang, D.J.; Legesse-Miller, A.; Johnson, E.L.; Coller, H.A. Regulation of the let-7a-3 Promoter by NF-κB. PLoS ONE 2012, 7, e31240. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An Epigenetic Switch Involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef]
- Kang, M.; Lee, K.-H.; Lee, H.S.; Jeong, C.W.; Ku, J.H.; Kim, H.H.; Kwak, C. Concurrent treatment with simvastatin and NF-κB inhibitor in human castration-resistant prostate cancer cells exerts synergistic anti-cancer effects via control of the NF-κB/LIN28/let-7 miRNA signaling pathway. PLoS ONE 2017, 12, e0184644. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, A.; Hakeem, A.R.; Radhakrishnan, S.; Reddy, M.S.; Rela, M. The Role and Therapeutic Potential of NF-kappa-B Pathway in Severe COVID-19 Patients. Inflammopharmacology 2020, 29, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Gustine, J.N.; Jones, D. Immunopathology of Hyperinflammation in COVID-19. Am. J. Pathol. 2020, 191, 4–17. [Google Scholar] [CrossRef]
- Coomes, E.A.; Haghbayan, H. Interleukin-6 in COVID-19: A systematic review and meta-analysis. Rev. Med. Virol. 2020, 30, e2141. [Google Scholar] [CrossRef] [PubMed]
- Brasier, A.R. The nuclear factor- B-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc. Res. 2010, 86, 211–218. [Google Scholar] [CrossRef]
- Gérard, C.; Gonze, D.; Lemaigre, F.; Novák, B. A Model for the Epigenetic Switch Linking Inflammation to Cell Transformation: Deterministic and Stochastic Approaches. PLOS Comput. Biol. 2014, 10, e1003455. [Google Scholar] [CrossRef]
- Tan, L.Y.; Komarasamy, T.V.; Balasubramaniam, V.R. Hyperinflammatory Immune Response and COVID-19: A Double Edged Sword. Front. Immunol. 2021, 12, 742941. [Google Scholar] [CrossRef]
- Niedźwiedzka-Rystwej, P.; Majchrzak, A.; Kurkowska, S.; Małkowska, P.; Sierawska, O.; Hrynkiewicz, R.; Parczewski, M. Immune Signature of COVID-19: In-Depth Reasons and Consequences of the Cytokine Storm. Int. J. Mol. Sci. 2022, 23, 4545. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).