Overlapping Gene Expression and Molecular Features in High-Grade B-Cell Lymphoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection
2.2. DNA/RNA Preparation
2.3. Gene Expression Analysis
2.4. Mutation Analysis
2.5. Analysis of IG Gene Rearrangements
3. Results
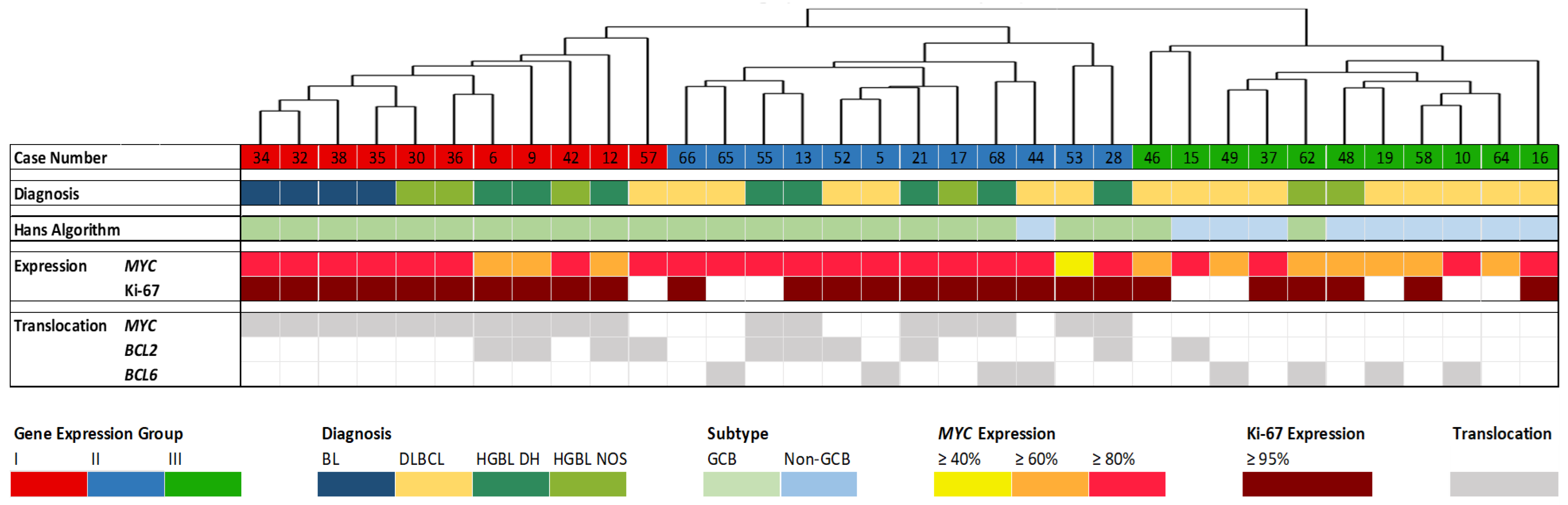
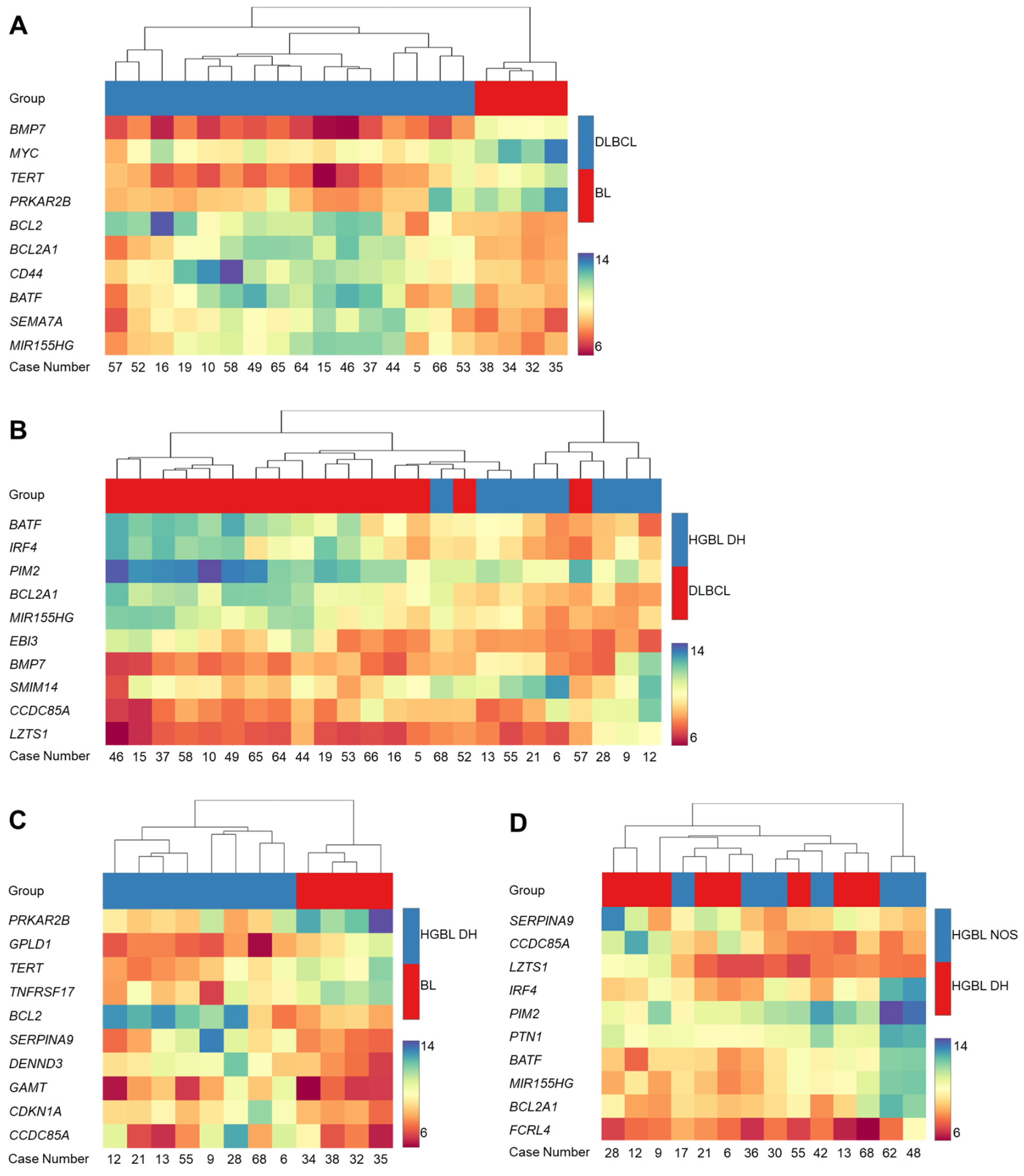
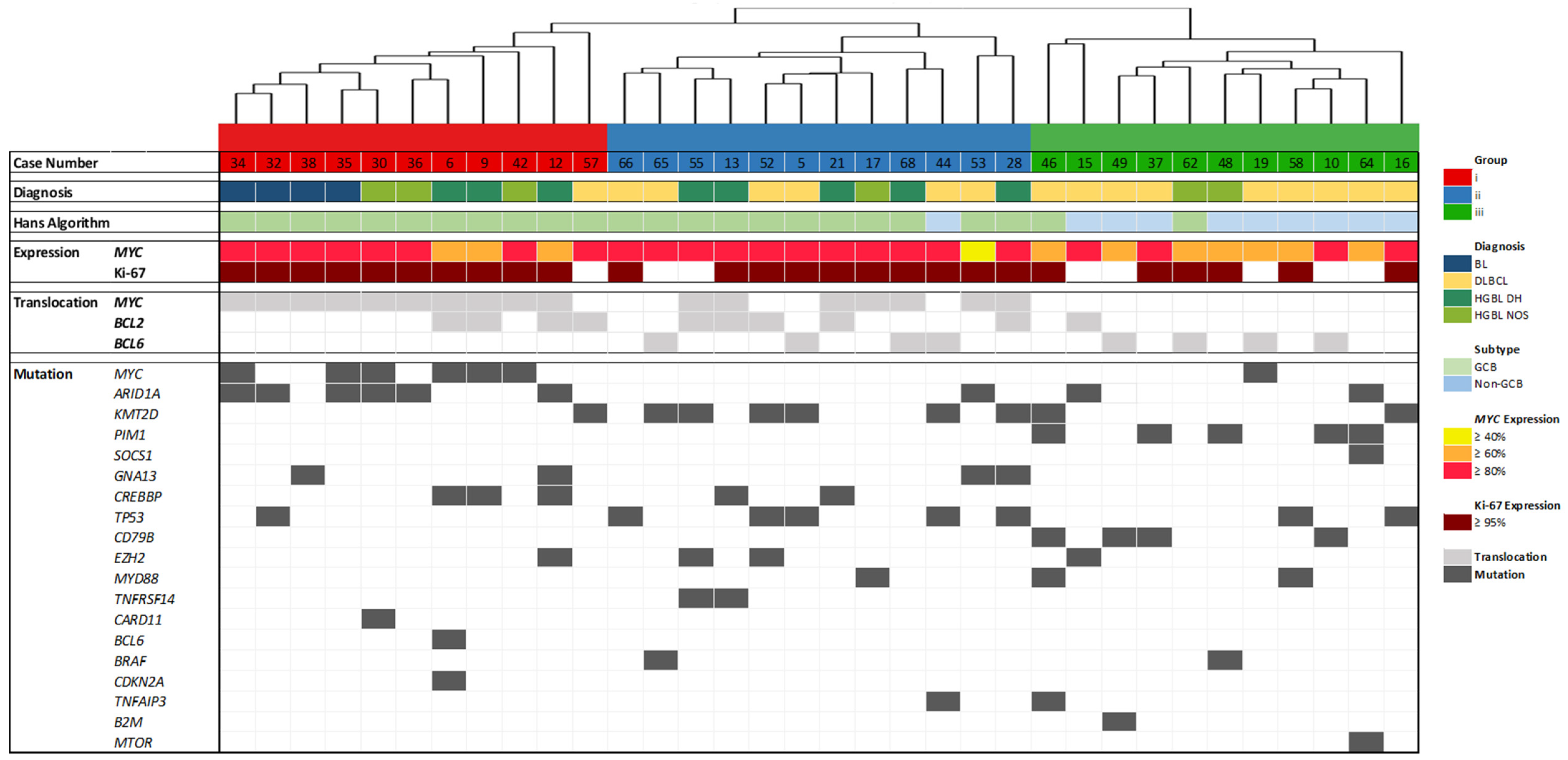
3.1. Gene Expression Analysis by HTG EdgeSeq
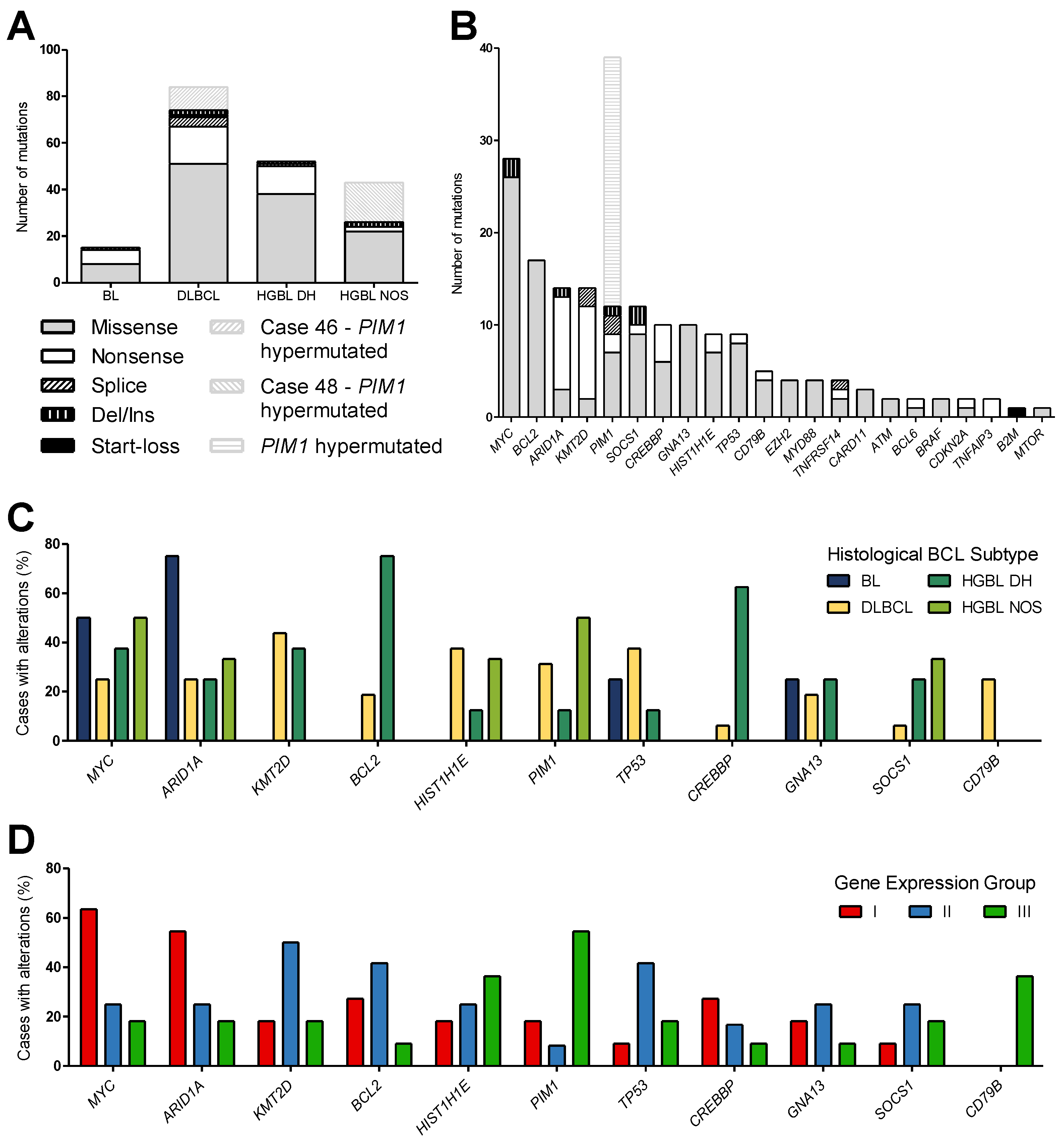
3.2. Mutation Analysis
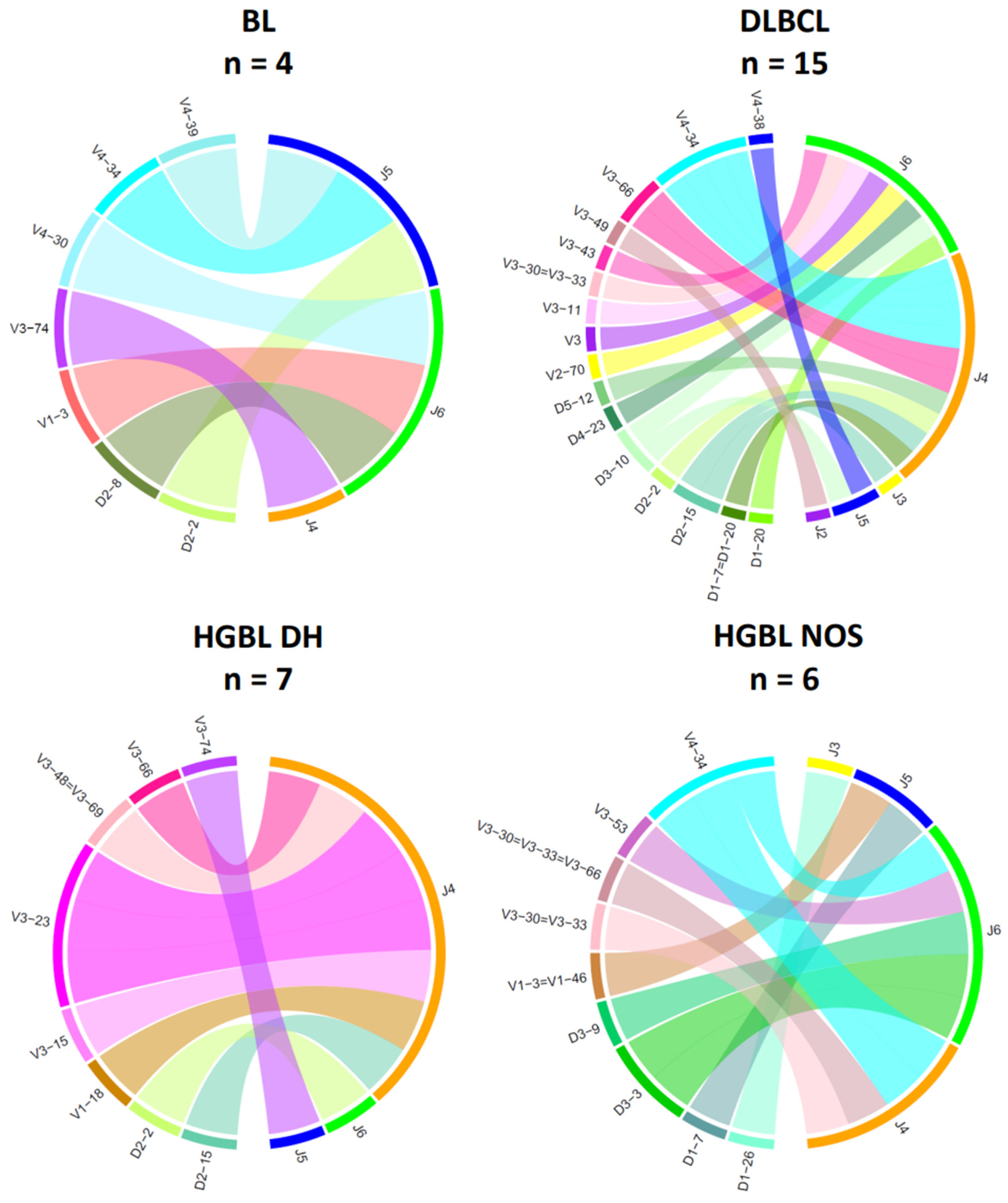
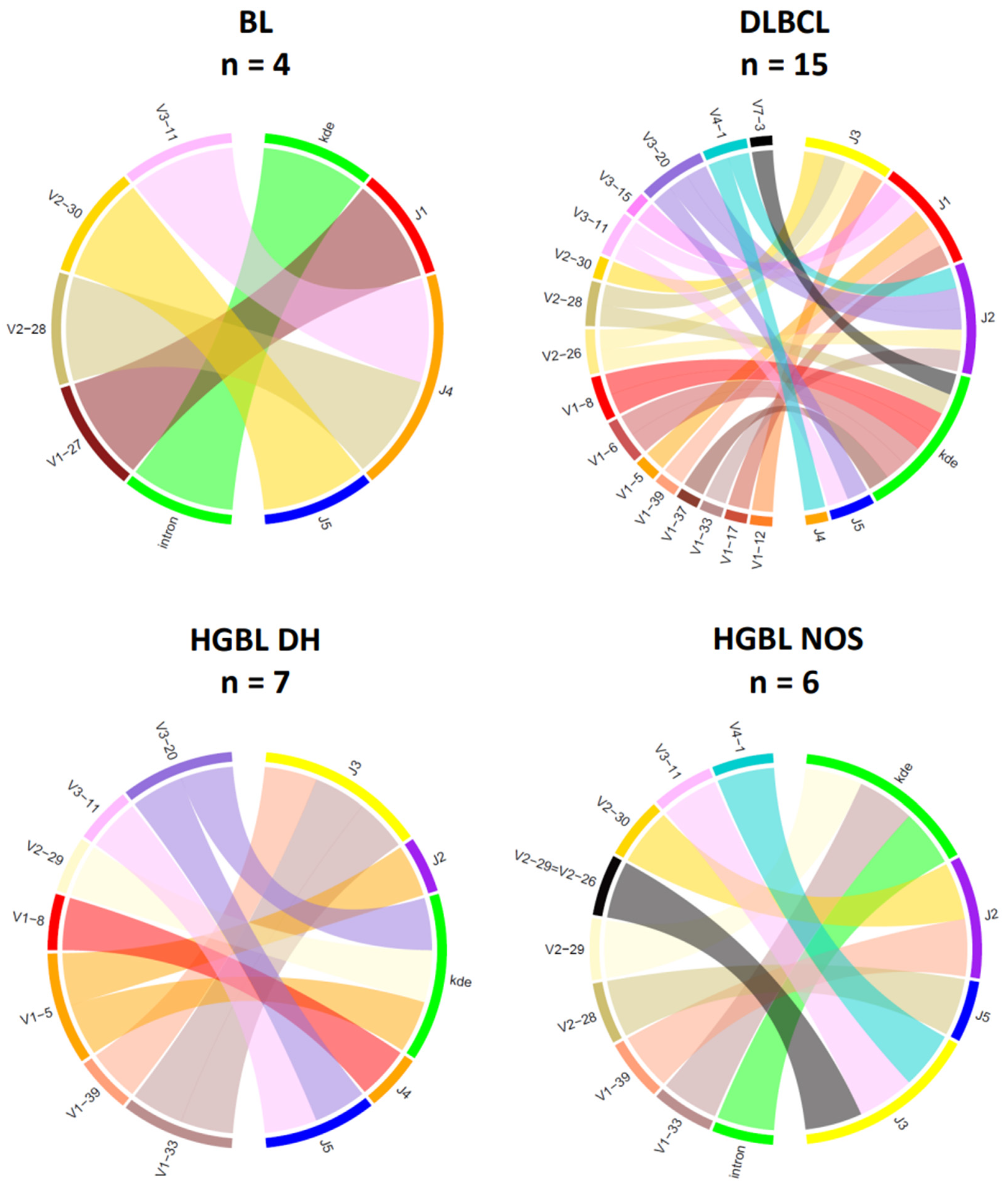
3.3. Analysis of IG Gene Rearrangements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fend, F.; Quintanilla-Martinez, L.; Kumar, S.; Beaty, M.W.; Blum, L.; Sorbara, L.; Jaffe, E.S.; Raffeld, M. Composite Low Grade B-Cell Lymphomas with Two Immunophenotypically Distinct Cell Populations Are True Biclonal Lymphomas. Am. J. Pathol. 1999, 154, 1857–1866. [Google Scholar] [CrossRef] [PubMed]
- Mugnaini, E.N.; Ghosh, N. Lymphoma. Prim. Care Clin. Off. Pract. 2016, 43, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.; Campo, E.; Harris, N.; Jaffe, E.; Pileri, S.; Stein, H.; Thiele, J.; Vardiman, J. WHO Classification: Tumours of the Haematopoietic and Lymphoid Tissues. IARC Press: Lyon, France, 2008; ISBN 9789283224310. [Google Scholar]
- Coupland, S.E.; Du, M.; Ferry, J.A.; de Jong, D.; Khoury, J.D.; Leoncini, L.; Naresh, K.N.; Ott, G.; Siebert, R.; Xerri, L. The Fifth Edition of the WHO Classification of Mature B-cell Neoplasms: Open Questions for Research. J. Pathol. 2024, 262, 255–270. [Google Scholar] [CrossRef]
- Dave, S.S.; Fu, K.; Wright, G.W.; Lam, L.T.; Kluin, P.; Boerma, E.-J.; Greiner, T.C.; Weisenburger, D.D.; Rosenwald, A.; Ott, G.; et al. Molecular Diagnosis of Burkitt’s Lymphoma. N. Engl. J. Med. 2006, 354, 2431–2442. [Google Scholar] [CrossRef]
- Hummel, M.; Bentink, S.; Berger, H.; Klapper, W.; Wessendorf, S.; Barth, T.F.E.; Bernd, H.-W.; Cogliatti, S.B.; Dierlamm, J.; Feller, A.C.; et al. A Biologic Definition of Burkitt’s Lymphoma from Transcriptional and Genomic Profiling. N. Engl. J. Med. 2006, 354, 2419–2430. [Google Scholar] [CrossRef]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human C-Myc Onc Gene Is Located on the Region of Chromosome 8 That Is Translocated in Burkitt Lymphoma Cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef]
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the C-Myc Gene into the Immunoglobulin Heavy Chain Locus in Human Burkitt Lymphoma and Murine Plasmacytoma Cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef]
- Karunakaran, P.; Selvarajan, G.; PK, J.; Mehra, N.; Sundersingh, S.; Dhanushkodi, M.; Rajan, A.K.; Kesana, S.; Sagar, T.G.; Kannan, K.; et al. High-Grade B-Cell Lymphoma,NOS: Does Prognosis End with the Diagnosis?—A Retrospective Analysis. Blood 2020, 136, 28–29. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Yao, Z.; Zhang, M. High-Grade B-Cell Lymphomas, Not Otherwise Specified: A Study of 41 Cases. Cancer Manag. Res. 2020, 12, 1903–1912. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.-M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating Morphologic, Clinical, and Genomic Data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef] [PubMed]
- Hilton, L.K.; Collinge, B.; Ben-Neriah, S.; Alduaij, W.; Shaalan, H.; Weng, A.P.; Cruz, M.; Slack, G.W.; Farinha, P.; Miyata-Takata, T.; et al. Motive and Opportunity: MYC Rearrangements in High-Grade B-Cell Lymphoma with MYC and BCL2 Rearrangements (an LLMPP Study). Blood 2024, 144, 525–540. [Google Scholar] [CrossRef]
- Yamashita, T.; Vollbrecht, C.; Hirsch, B.; Kleo, K.; Anagnostopoulos, I.; Hummel, M. Integrative Genomic Analysis Focused on Cell Cycle Genes for MYC-Driven Aggressive Mature B-Cell Lymphoma. J. Clin. Exp. Hematop. 2020, 60, 87–96. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. DbSNP: The NCBI Database of Genetic Variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Henrie, A.; Hemphill, S.E.; Ruiz-Schultz, N.; Cushman, B.; DiStefano, M.T.; Azzariti, D.; Harrison, S.M.; Rehm, H.L.; Eilbeck, K. ClinVar Miner: Demonstrating Utility of a Web-Based Tool for Viewing and Filtering ClinVar Data. Hum. Mutat. 2018, 39, 1051–1060. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- van den Brand, M.; Rijntjes, J.; Möbs, M.; Steinhilber, J.; van der Klift, M.Y.; Heezen, K.C.; Kroeze, L.I.; Reigl, T.; Porc, J.; Darzentas, N.; et al. Next-Generation Sequencing-Based Clonality Assessment of Ig Gene Rearrangements: A Multicenter Validation Study by EuroClonality-NGS. J. Mol. Diagn. 2021, 23, 1105–1115. [Google Scholar] [CrossRef]
- Scheijen, B.; Meijers, R.W.J.; Rijntjes, J.; van der Klift, M.Y.; Möbs, M.; Steinhilber, J.; Reigl, T.; van den Brand, M.; Kotrová, M.; Ritter, J.-M.; et al. Next-Generation Sequencing of Immunoglobulin Gene Rearrangements for Clonality Assessment: A Technical Feasibility Study by EuroClonality-NGS. Leukemia 2019, 33, 2227–2240. [Google Scholar] [CrossRef]
- Bystry, V.; Reigl, T.; Krejci, A.; Demko, M.; Hanakova, B.; Grioni, A.; Knecht, H.; Schlitt, M.; Dreger, P.; Sellner, L.; et al. ARResT/Interrogate: An Interactive Immunoprofiler for IG/TR NGS Data. Bioinformatics 2017, 33, 435–437. [Google Scholar] [CrossRef] [PubMed]
- Hans, C.P. Confirmation of the Molecular Classification of Diffuse Large B-Cell Lymphoma by Immunohistochemistry Using a Tissue Microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Zeisberg, M.; Hanai, J.; Sugimoto, H.; Mammoto, T.; Charytan, D.; Strutz, F.; Kalluri, R. BMP-7 Counteracts TGF-Β1–Induced Epithelial-to-Mesenchymal Transition and Reverses Chronic Renal Injury. Nat. Med. 2003, 9, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Huse, K.; Bakkebø, M.; Oksvold, M.P.; Forfang, L.; Hilden, V.I.; Stokke, T.; Smeland, E.B.; Myklebust, J.H. Bone Morphogenetic Proteins Inhibit CD40L/IL-21-Induced Ig Production in Human Bcells: Differential Effects of BMP-6 and BMP-7. Eur. J. Immunol. 2011, 41, 3135–3145. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.S.; Kim, C.; Cheng, C.Y.; Brown, S.H.J.; Wu, J.; Kannan, N. Signaling through CAMP and CAMP-Dependent Protein Kinase: Diverse Strategies for Drug Design. Biochim. Biophys. Acta 2008, 1784, 16–26. [Google Scholar] [CrossRef]
- Vogler, M. BCL2A1: The Underdog in the BCL2 Family. Cell Death Differ. 2012, 19, 67–74. [Google Scholar] [CrossRef]
- Wu, K.-J.; Grandori, C.; Amacker, M.; Simon-Vermot, N.; Polack, A.; Lingner, J.; Dalla-Favera, R. Direct Activation of TERT Transcription by C-MYC. Nat. Genet. 1999, 21, 220–224. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Gall, J.G. A Tandemly Repeated Sequence at the Termini of the Extrachromosomal Ribosomal RNA Genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A Highly Conserved Repetitive DNA Sequence, (TTAGGG)n, Present at the Telomeres of Human Chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- López, C.; Kleinheinz, K.; Aukema, S.M.; Rohde, M.; Bernhart, S.H.; Hübschmann, D.; Wagener, R.; Toprak, U.H.; Raimondi, F.; Kreuz, M.; et al. Genomic and Transcriptomic Changes Complement Each Other in the Pathogenesis of Sporadic Burkitt Lymphoma. Nat. Commun. 2019, 10, 1459. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.D.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The C-Myc Target Gene Network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Cole, M.D. Mechanism of Transcriptional Activation by the Myc Oncoproteins. Semin. Cancer Biol. 2006, 16, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. C-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Blomme, S.; De Paepe, P.; Devos, H.; Emmerechts, J.; Snauwaert, S.; Cauwelier, B. Alternative Genetic Alterations of MYC, BCL2, and/or BCL6 in High-grade B-cell Lymphoma (HGBL) and Diffuse Large B-cell Lymphoma (DLBCL): Can We Identify Different Prognostic Subgroups? Genes Chromosomes Cancer 2024, 63, e23211. [Google Scholar] [CrossRef]
- Menssen, A.; Hermeking, H. Characterization of the C-MYC-Regulated Transcriptome by SAGE: Identification and Analysis of c-MYC Target Genes. Proc. Natl. Acad. Sci. USA 2002, 99, 6274–6279. [Google Scholar] [CrossRef]
- Feuerhake, F. NF B Activity, Function, and Target-Gene Signatures in Primary Mediastinal Large B-Cell Lymphoma and Diffuse Large B-Cell Lymphoma Subtypes. Blood 2005, 106, 1392–1399. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Author Correction: Molecular Subtypes of Diffuse Large B Cell Lymphoma Are Associated with Distinct Pathogenic Mechanisms and Outcomes. Nat. Med. 2018, 24, 1290–1291. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-KappaB Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sundaram, C.; Reuter, S.; Aggarwal, B.B. Inhibiting NF-ΚB Activation by Small Molecules as a Therapeutic Strategy. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2010, 1799, 775–787. [Google Scholar] [CrossRef]
- Grumont, R.J.; Rourke, I.J.; Gerondakis, S. Rel-Dependent Induction of A1 Transcription Is Required to Protect B Cells from Antigen Receptor Ligation-Induced Apoptosis. Genes Dev. 1999, 13, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Hinz, M.; Lemke, P.; Anagnostopoulos, I.; Hacker, C.; Krappmann, D.; Mathas, S.; Doerken, B.; Zenke, M.; Stein, H.; Scheidereit, C. Nuclear Factor ΚB–Dependent Gene Expression Profiling of Hodgkin’s Disease Tumor Cells, Pathogenetic Significance, and Link to Constitutive Signal Transducer and Activator of Transcription 5a Activity. J. Exp. Med. 2002, 196, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ohtani, K.; Iwanaga, R.; Matsumura, Y.; Nakamura, M. Direct Trans-Activation of the Human Cyclin D2 Gene by the Oncogene Product Tax of Human T-Cell Leukemia Virus Type I. Oncogene 2001, 20, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.-J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of MiR-155 and MiR-125b Levels Following Lipopolysaccharide/TNF-α Stimulation and Their Possible Roles in Regulating the Response to Endotoxin Shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.-J.; van den Berg, A. BIC and MiR-155 Are Highly Expressed in Hodgkin, Primary Mediastinal and Diffuse Large B Cell Lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J. Risk of Regorafenib-Induced Cardiovascular Events in Patients with Solid Tumors. Medicine 2018, 97, e12705. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Echlin, D.R.; Tae, H.-J.; Mitin, N.; Taparowsky, E.J. B-ATF Functions as a Negative Regulator of AP-1 Mediated Transcription and Blocks Cellular Transformation by Ras and Fos. Oncogene 2000, 19, 1752–1763. [Google Scholar] [CrossRef]
- Niedobitek, G.; Päzolt, D.; Teichmann, M.; Devergne, O. Frequent Expression of the Epstein-Barr Virus (EBV)-Induced Gene, EBI3, an IL-12 P40-Related Cytokine, in Hodgkin and Reed-Sternberg Cells. J. Pathol. 2002, 198, 310–316. [Google Scholar] [CrossRef]
- Gonin, J.; Larousserie, F.; Bastard, C.; Picquenot, J.-M.; Couturier, J.; Radford-Weiss, I.; Dietrich, C.; Brousse, N.; Vacher-Lavenu, M.-C.; Devergne, O. Epstein-Barr Virus-Induced Gene 3 (EBI3): A Novel Diagnosis Marker in Burkitt Lymphoma and Diffuse Large B-Cell Lymphoma. PLoS ONE 2011, 6, e24617. [Google Scholar] [CrossRef]
- Gómez-Abad, C.; Pisonero, H.; Blanco-Aparicio, C.; Roncador, G.; González-Menchén, A.; Martinez-Climent, J.A.; Mata, E.; Rodríguez, M.E.; Muñoz-González, G.; Sánchez-Beato, M.; et al. PIM2 Inhibition as a Rational Therapeutic Approach in B-Cell Lymphoma. Blood 2011, 118, 5517–5527. [Google Scholar] [CrossRef] [PubMed]
- Panea, R.I.; Love, C.L.; Shingleton, J.R.; Reddy, A.; Bailey, J.A.; Moormann, A.M.; Otieno, J.A.; Ong’echa, J.M.; Oduor, C.I.; Schroeder, K.M.S.; et al. The Whole-Genome Landscape of Burkitt Lymphoma Subtypes. Blood 2019, 134, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Jun, M.-H.; Jun, Y.-W.; Kim, K.-H.; Lee, J.-A.; Jang, D.-J. Characterization of the Cellular Localization of C4orf34 as a Novel Endoplasmic Reticulum Resident Protein. BMB Rep. 2014, 47, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qiu, L.; Xu, J.; Lin, P.; Ok, C.Y.; Tang, G.; McDonnell, T.J.; James You, M.; Khanlari, M.; Miranda, R.N.; et al. High-Grade B-Cell Lymphoma (HGBL)-NOS Is Clinicopathologically and Genetically More Similar to DLBCL/HGBL-DH than DLBCL. Leukemia 2023, 37, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Pasqualucci, L.; Dalla-Favera, R. Genetics of Diffuse Large B-Cell Lymphoma. Blood 2018, 131, 2307–2319. [Google Scholar] [CrossRef]
- Haberl, S.; Haferlach, T.; Stengel, A.; Jeromin, S.; Kern, W.; Haferlach, C. MYC Rearranged B-Cell Neoplasms: Impact of Genetics on Classification. Cancer Genet. 2016, 209, 431–439. [Google Scholar] [CrossRef]
- Cucco, F.; Barrans, S.; Sha, C.; Clipson, A.; Crouch, S.; Dobson, R.; Chen, Z.; Thompson, J.S.; Care, M.A.; Cummin, T.; et al. Distinct Genetic Changes Reveal Evolutionary History and Heterogeneous Molecular Grade of DLBCL with MYC/BCL2 Double-Hit. Leukemia 2020, 34, 1329–1341. [Google Scholar] [CrossRef]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-Wide Discovery of Somatic Coding and Noncoding Mutations in Pediatric Endemic and Sporadic Burkitt Lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef]
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.W.L.; Srivastava, G.; et al. The Genetic Landscape of Mutations in Burkitt Lymphoma. Nat. Genet. 2012, 44, 1321–1325. [Google Scholar] [CrossRef]
- Reddy, A.; Zhang, J.; Davis, N.S.; Moffitt, A.B.; Love, C.L.; Waldrop, A.; Leppa, S.; Pasanen, A.; Meriranta, L.; Karjalainen-Lindsberg, M.-L.; et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017, 171, 481–494.e15. [Google Scholar] [CrossRef]
- Fan, Z.; Pei, R.; Sha, K.; Chen, L.; Wang, T.; Lu, Y. Comprehensive Characterization of Driver Genes in Diffuse Large B Cell Lymphoma. Oncol. Lett. 2020, 20, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.E.; Brown, K.D.; Siebenlist, U.; Staudt, L.M. Constitutive Nuclear Factor ΚB Activity Is Required for Survival of Activated B Cell–like Diffuse Large B Cell Lymphoma Cells. J. Exp. Med. 2001, 194, 1861–1874. [Google Scholar] [CrossRef] [PubMed]
- Compagno, M.; Lim, W.K.; Grunn, A.; Nandula, S.V.; Brahmachary, M.; Shen, Q.; Bertoni, F.; Ponzoni, M.; Scandurra, M.; Califano, A.; et al. Mutations of Multiple Genes Cause Deregulation of NF-ΚB in Diffuse Large B-Cell Lymphoma. Nature 2009, 459, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Young, R.M.; Ceribelli, M.; Jhavar, S.; Xiao, W.; Zhang, M.; Wright, G.; Shaffer, A.L.; Hodson, D.J.; Buras, E.; et al. Burkitt Lymphoma Pathogenesis and Therapeutic Targets from Structural and Functional Genomics. Nature 2012, 490, 116–120. [Google Scholar] [CrossRef]
- Bouska, A.; Bi, C.; Lone, W.; Zhang, W.; Kedwaii, A.; Heavican, T.; Lachel, C.M.; Yu, J.; Ferro, R.; Eldorghamy, N.; et al. Adult High-Grade B-Cell Lymphoma with Burkitt Lymphoma Signature: Genomic Features and Potential Therapeutic Targets. Blood 2017, 130, 1819–1831. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Künstner, A.; Witte, H.M.; Riedl, J.; Bernard, V.; Stölting, S.; Merz, H.; Olschewski, V.; Peter, W.; Ketzer, J.; Busch, Y.; et al. Mutational Landscape of High-Grade B-Cell Lymphoma with MYC-, BCL2 and/or BCL6 Rearrangements Characterized by Whole-Exome Sequencing. Haematologica 2022, 107, 1850–1863. [Google Scholar] [CrossRef]
| Gene | Transcript ID | BL | DLBCL | HGBL DH | HGBL NOS |
|---|---|---|---|---|---|
| ARID1A | NM_006015 | Q487*; R693*; Q1145Rfs*16; P1898Hfs*25 2 | A345_A349del; Q564*; P887L; Y1233H; P1898Hfs*25 | R1528*; R2116T | Q74*; Q1145Rfs*16 |
| ATM | NM_000051 | - | K2810Q | - | C1811Y |
| B2M | NM_004048 | - | M1V | - | - |
| BCL2 | NM_000657 | - | H3L; H58R; A67G; P78L; V92A; H120E; F124C | G5A; N11T; R12W; D31G; G47D; Q52L; A60D; P90S; N182D; G197S | - |
| BCL6 | NM_001706 | - | R526C | Q397* | - |
| BRAF | NM_004333 | - | G469E | - | G466E |
| CARD11 | NM_032415 | - | R235P; G757V | - | R235P |
| CD79B | NM_001039933 | - | M191L; Y197C; Y197H; Y197S; Y208* | - | - |
| CDKN2A | NM_000077 | - | V82M | R80* | - |
| CREBBP | NM_004380 | - | Y1503F | Q1073Vfs*13; R1173*; V1371D; S1436R; R1446H; P1494R; R1498Q; D1543V | - |
| EZH2 | NM_004456 | - | Y646F; Y646N | Y646F 2 | - |
| GNA13 | NM_006572 | Q27*; H345R | V221L; N263K; L342_F346delins* | Q28*; N91D; L184P; R260*; V362M | - |
| HIST1H1E | NM_005321 | - | P118S 2; A123T 2; T142S; A167G; P201A | G103A | G103R; A165V |
| KMT2D | NM_003482 | - | c.15784+1G>A; c.4693+6T>A; L1199Hfs*7; Q1893*; R2771*; I3421Mfs*12; P3466Tfs*2; Q3609*; R5048C | Q2416*; R2801*; R2847H; Q4732*; Q5003* | - |
| MTOR | NM_004958 | - | W1456R | - | - |
| MYC | NM_002467 | Q50H; E54D; T73S; F153C; L191F; S217_S218delinsIN; S264R | P72S; T73A; P75T; Q194H | Q10H; Q51_E54delinsHQID; S53I; P72S; T73I; V92I; K158N; N451K | S21N; N26T; E54D 2; T73I; S107R; E137D; G152A; S187C |
| MYD88 | NM_002468 | - | S219C; L265P 2 | - | L265P |
| PIM1 | NM_001243186 | - | c.513+1G>A; c.880+1G>A; Q30L; G73D; Y129*; Q130*; V217M; L273F; [c.880+5G>A; Q128H; P172S; P178S; L184V; S188R; Q218K; I224Gfs*74; L265F; L323V]Case 46 | N251I | I147_S150del; E161Q; S188T; [c.514-1_521 delinsACCTAATGA; V90F; G91E; G119_P124delins DKKKEA; P124A; G139D; M179I; S188N; L197F; V217_D219delinsLQH; Q218*; E226K; Q231*; V242I; L275F; R312K; C361Y]Case 48 |
| SOCS1 | NM_003745 | - | A16S; Y64Afs*48; S116_V117delinsRL | E24D; Y64N; C111W | A3T; A3V; R59H; D138H; Y154_R170del; M161I |
| TNFAIP3 | NM_001270508 | - | C103*; F152Lfs*64 | - | - |
| TNFRSF14 | NM_003820 | - | V239I | c.50_69+1delinsGAACCGACGT; Y61*; N110S | - |
| TP53 | NM_000546 | D281E | A138V; R181P; R196*; Y234C; R273H; R273G | K139N; Y234D | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faißt, K.D.; Husemann, C.C.; Kleo, K.; Twardziok, M.; Hummel, M. Overlapping Gene Expression and Molecular Features in High-Grade B-Cell Lymphoma. J. Mol. Pathol. 2024, 5, 415-436. https://doi.org/10.3390/jmp5040028
Faißt KD, Husemann CC, Kleo K, Twardziok M, Hummel M. Overlapping Gene Expression and Molecular Features in High-Grade B-Cell Lymphoma. Journal of Molecular Pathology. 2024; 5(4):415-436. https://doi.org/10.3390/jmp5040028
Chicago/Turabian StyleFaißt, Katharina D., Cora C. Husemann, Karsten Kleo, Monika Twardziok, and Michael Hummel. 2024. "Overlapping Gene Expression and Molecular Features in High-Grade B-Cell Lymphoma" Journal of Molecular Pathology 5, no. 4: 415-436. https://doi.org/10.3390/jmp5040028
APA StyleFaißt, K. D., Husemann, C. C., Kleo, K., Twardziok, M., & Hummel, M. (2024). Overlapping Gene Expression and Molecular Features in High-Grade B-Cell Lymphoma. Journal of Molecular Pathology, 5(4), 415-436. https://doi.org/10.3390/jmp5040028






