MET Exon 14 Variants in Non-Small Cell Lung Carcinoma: Prevalence, Clinicopathologic and Molecular Features
Abstract
1. Introduction
2. Subjects and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fujino, T.; Suda, K.; Mitsudomi, T. Lung Cancer with MET exon 14 Skipping Mutation: Genetic Feature, Current Treatments, and Future Challenges. Lung Cancer: Targets Ther. 2021, 12, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zou, Q.; Liu, H.; Qiu, B.; Li, Q.; Lin, Y.; Liang, Y. Management of Non-small Cell Lung Cancer Patients with MET Exon 14 Skipping Mutations. Curr. Treat. Options Oncol. 2020, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Ho, A.T.N.; Altibi, A.M.; Nakazawa, T.; Katoh, R.; Kondo, T. Clinicopathological implications of MET exon 14 mutations in non-small cell lung cancer—A systematic review and meta-analysis. Lung Cancer 2018, 123, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET Exon 14 Mutations in Non–Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef]
- Schrock, A.B.; Frampton, G.M.; Suh, J.; Chalmers, Z.R.; Rosenzweig, M.; Erlich, R.L.; Halmos, B.; Goldman, J.; Forde, P.; Leuenberger, K.; et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J. Thorac. Oncol. 2016, 11, 1493–1502. [Google Scholar] [CrossRef]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.H.; Chung, L.Y.; Chau, S.L.; Lung, R.W.M.; Tong, C.Y.; Chow, C.; Tin, E.K.Y.; Yu, Y.H.; et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non–Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef]
- Ou, S.-H.I.; Kwak, E.L.; Siwak-Tapp, C.; Dy, J.; Bergethon, K.; Clark, J.W.; Camidge, D.R.; Solomon, B.J.; Maki, R.G.; Bang, Y.-J.; et al. Activity of Crizotinib (PF02341066), a Dual Mesenchymal-Epithelial Transition (MET) and Anaplastic Lymphoma Kinase (ALK) Inhibitor, in a Non-small Cell Lung Cancer Patient with De Novo MET Amplification. J. Thorac. Oncol. 2011, 6, 942–946. [Google Scholar] [CrossRef]
- Jørgensen, J.T.; Mollerup, J. Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC. Cancers 2022, 14, 2150. [Google Scholar] [CrossRef]
- Lung Cancer Statistics. Available online: https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html (accessed on 14 February 2021).
- Salgia, R.; Sattler, M.; Scheele, J.; Stroh, C.; Felip, E. The promise of selective MET inhibitors in non-small cell lung cancer with MET exon 14 skipping. Cancer Treat. Rev. 2020, 87, 102022. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, R.; Ye, T.; Yu, S.; Hu, H.; Shen, X.; Li, Y.; Ji, H.; Sun, Y.; Chen, H. MET exon 14 skipping defines a unique molecular class of non-small cell lung cancer. Oncotarget 2016, 7, 41691–41702. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Gou, L.-Y.; Li, A.-N.; Lou, N.-N.; Gao, H.-F.; Su, J.; Yang, J.-J.; Zhang, X.-C.; Shao, Y.; Dong, Z.-Y.; et al. The Unique Characteristics of MET Exon 14 Mutation in Chinese Patients with NSCLC. J. Thorac. Oncol. 2016, 11, 1503–1510. [Google Scholar] [CrossRef]
- Poirot, B.; Doucet, L.; Benhenda, S.; Champ, J.; Meignin, V.; Lehmann-Che, J. MET Exon 14 Alterations and New Resistance Mutations to Tyrosine Kinase Inhibitors: Risk of Inadequate Detection with Current Amplicon-Based NGS Panels. J. Thorac. Oncol. 2017, 12, 1582–1587. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.D.; Lomboy, A.; Lawrence, C.A.; Yourshaw, M.; Bocsi, G.T.; Camidge, D.R.; Aisner, D.L. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. J. Thorac. Oncol. 2019, 14, 737–741. [Google Scholar] [CrossRef]
- Wang, S.X.; Zhang, B.M.; Wakelee, H.A.; Koontz, M.Z.; Pan, M.; Diehn, M.; Kunder, C.A.; Neal, J.W. Case series of MET exon 14 skipping mutation-positive non-small-cell lung cancers with response to crizotinib and cabozantinib. Anti-Cancer Drugs 2019, 30, 537–541. [Google Scholar] [CrossRef]
- Saffroy, R.; Fallet, V.; Girard, N.; Mazieres, J.; Sibilot, D.M.; Lantuejoul, S.; Rouquette, I.; Thivolet-Bejui, F.; Vieira, T.; Antoine, M.; et al. MET exon 14 mutations as targets in routine molecular analysis of primary sarcomatoid carcinoma of the lung. Oncotarget 2017, 8, 42428–42437. [Google Scholar] [CrossRef] [PubMed]
- Filosso, P.L.; Ruffini, E.; Asioli, S.; Giobbe, R.; Macri, L.; Bruna, M.C.; Sandri, A.; Oliaro, A. Adenosquamous lung carcinomas: A histologic subtype with poor prognosis. Lung Cancer 2011, 74, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Maneenil, K.; Xue, Z.; Liu, M.; Boland, J.; Wu, F.; Stoddard, S.M.; Molina, J.; Yang, P. Sarcomatoid Carcinoma of the Lung: The Mayo Clinic Experience in 127 Patients. Clin. Lung Cancer 2018, 19, e323–e333. [Google Scholar] [CrossRef]
- Socinski, M.A.; Pennell, N.A.; Davies, K.D. METExon 14 Skipping Mutations in Non–Small-Cell Lung Cancer: An Overview of Biology, Clinical Outcomes, and Testing Considerations. JCO Precis. Oncol. 2021, 5, 653–663. [Google Scholar] [CrossRef]
- Awad, M.M.; Lee, J.K.; Madison, R.; Classon, A.; Kmak, J.; Frampton, G.M.; Alexander, B.M.; Venstrom, J.; Schrock, A.B. Characterization of 1,387 NSCLCs with MET exon 14 (METex14) skipping alterations (SA) and potential acquired resistance (AR) mechanisms. J. Clin. Oncol. 2020, 38, 9511. [Google Scholar] [CrossRef]
- Cohen, D.; Hondelink, L.M.; Solleveld-Westerink, N.; Uljee, S.M.; Ruano, D.; Cleton-Jansen, A.-M.; von der Thüsen, J.H.; Ramai, S.R.S.; Postmus, P.E.; van Roggen, J.F.G.; et al. Optimizing Mutation and Fusion Detection in NSCLC by Sequential DNA and RNA Sequencing. J. Thorac. Oncol. 2020, 15, 1000–1014. [Google Scholar] [CrossRef]
- Sabari, J.; Leonardi, G.; Shu, C.; Umeton, R.; Montecalvo, J.; Ni, A.; Chen, R.; Dienstag, J.; Mrad, C.; Bergagnini, I.; et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann. Oncol. 2018, 29, 2085–2091. [Google Scholar] [CrossRef]
- Xu, Z.; Li, H.; Dong, Y.; Cheng, P.; Luo, F.; Fu, S.; Gao, M.; Kong, L.; Che, N. Incidence and PD-L1 Expression of MET 14 Skipping in Chinese Population: A Non-Selective NSCLC Cohort Study Using RNA-Based Sequencing. OncoTargets Ther. 2020, 13, 6245–6253. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.A.; Weiss, J. Advances in the Treatment of Non–Small Cell Lung Cancer. Clin. Chest Med. 2020, 41, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Hellmann, M.D.; Rizvi, N.A.; Carcereny, E.; Leighl, N.B.; Ahn, M.-J.; Eder, J.P.; Balmanoukian, A.S.; Aggarwal, C.; Horn, L.; et al. Five-Year Overall Survival for Patients With Advanced Non-Small-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J. Clin. Oncol. 2019, 37, 2518–2527. [Google Scholar] [CrossRef] [PubMed]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Kurata, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018, 379, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Imyanitov, E.N.; Iyevleva, A.G.; Levchenko, E.V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol. 2021, 157, 103194. [Google Scholar] [CrossRef] [PubMed]
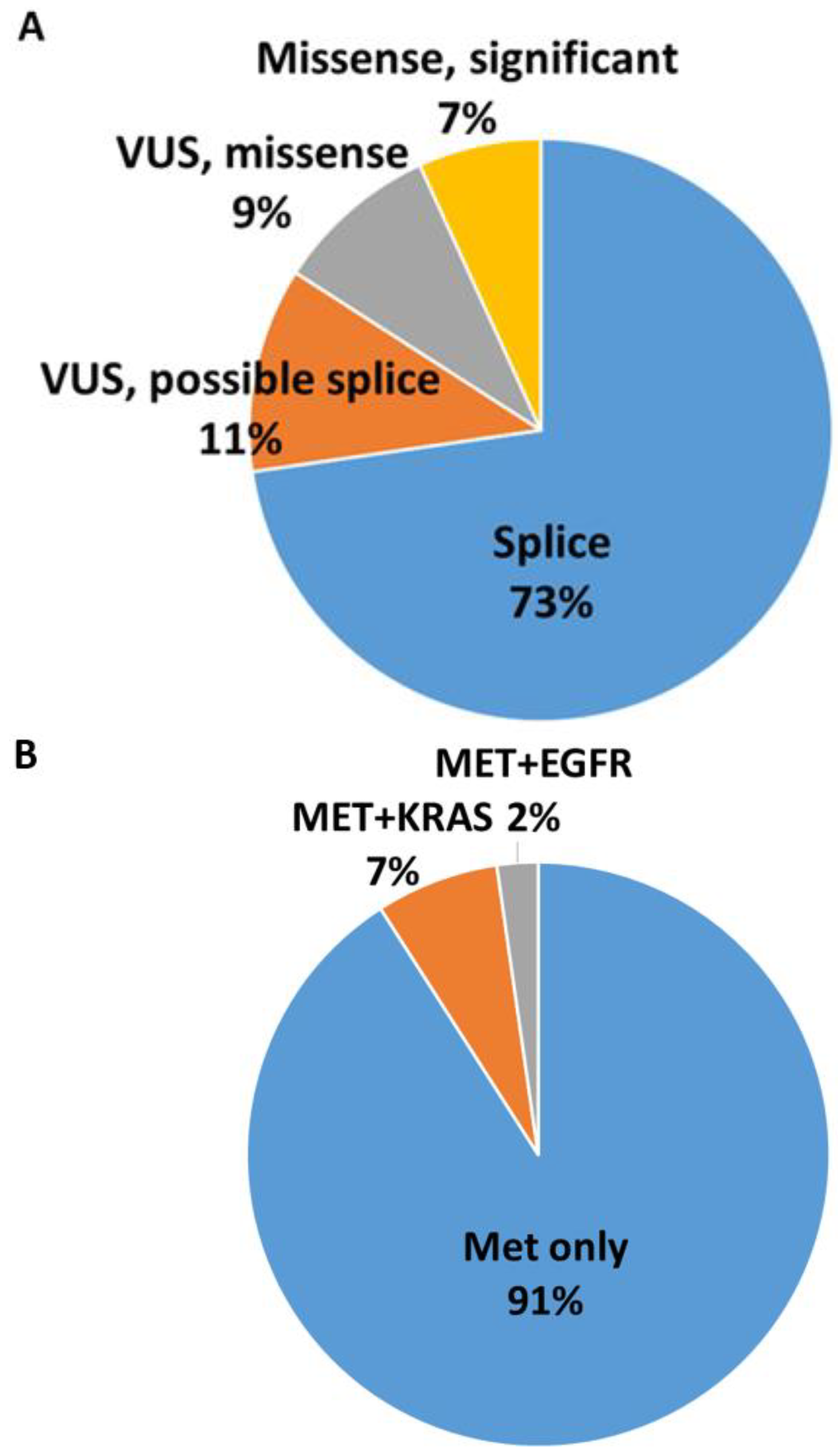
| Prevalence of MET Exon 14 Mutations | 1.9% | |||||
| Total patients | 44 patients: 26 men, 18 women | |||||
| Mean age: 76 years | ||||||
| Clinical stage available | Total 35 cases | |||||
| Stage 1 and 2: 20 cases (57%) | Stage 3: 3 cases (9%) | Stage 4: 12 cases (34%) | ||||
| Tumor resected | Total 19 cases | |||||
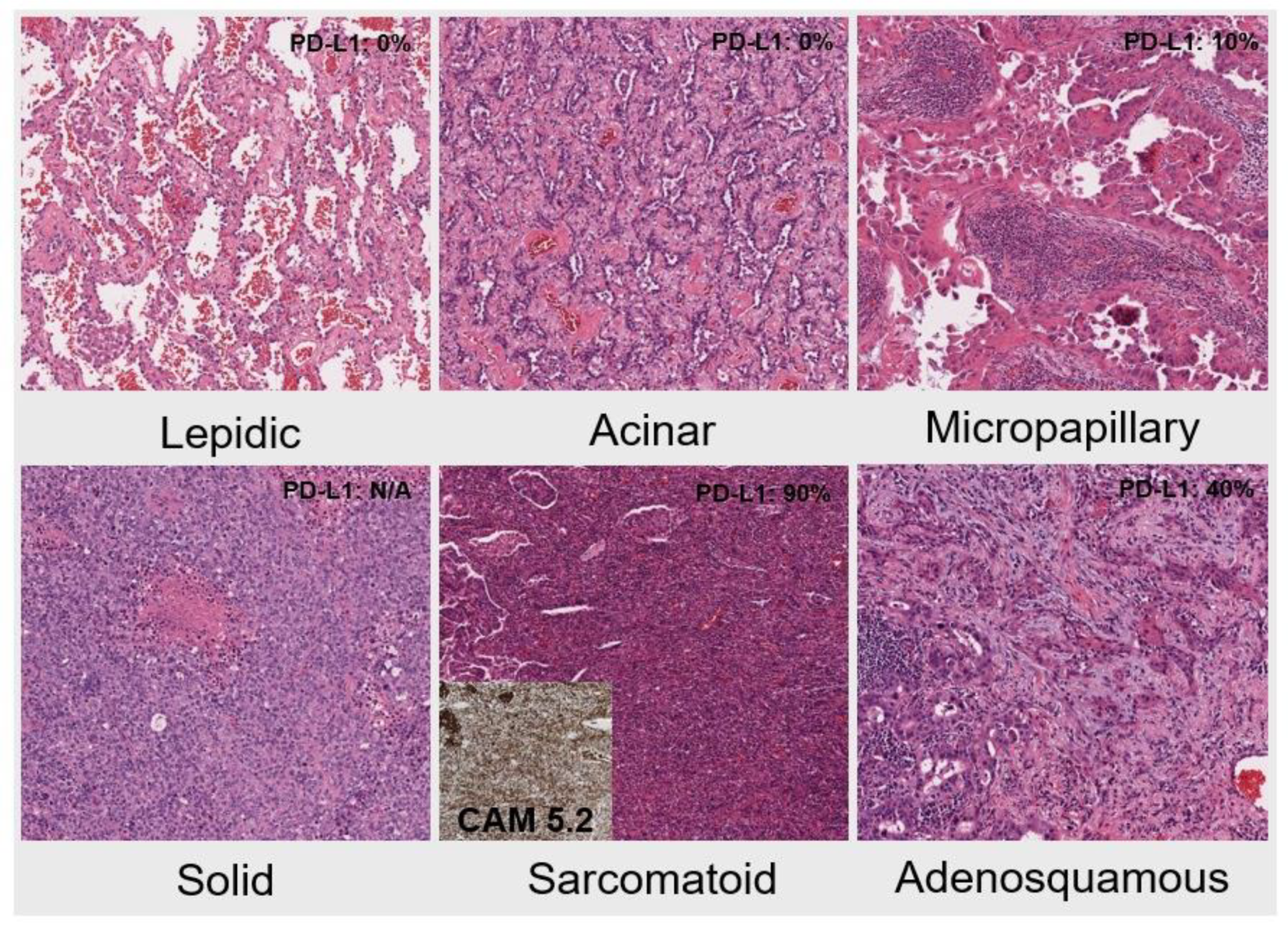
| Histologic type and growth pattern | ||||||
| Lepidic pattern-predominant | Acinar pattern-predominant | Micropapillary-predominant | Solid-predominant | Adeno-squamous | Sarcomatoid | |
| 7 | 6 | 2 | 1 | 2 | 1 | |
| PD-L1 expression | Total 27 cases, positive in 22 cases (82%) | |||||
| 0% | 1–49%: | >50% | ||||
| 5 cases (18%) | 11 cases (41%) | 11 cases (41%) | ||||
| Stage 1–3 | Stage 4 | |||||
| PD-L1 < 50% | PD-L1 > 50% | PD-L1 < 50% | PD-L1 > 50% | |||
| 12 cases (67%) | 6 cases (33%) | 2 cases (33%) | 4 cases (67%) | |||
| Case # | Result | AF | Significance | Type of MET Mutation | Exon14 Skipping | Other Mutations | Surgical Pathology Diagnosis on Resection |
|---|---|---|---|---|---|---|---|
| 1 | c.3028+2T>A | 41 | significant | splice | yes | Adenocarcinoma, lepidic predominant, with additional acinar component | |
| 2 | c.3280C>T (p.His1094Tyr) | 11 | significant | missense | no | KRAS | |
| 3 | c.3028+2del | 11 | significant | splice | yes | Adenocarcinoma, acinar predominant (70%) with lepidic pattern (30%). | |
| 4 | c.3028G>C (p.Asp1010His) | 3 | significant | splice | yes | ||
| 5 | c.3023_3028+7delinsTC | 91 | significant | splice | yes | ||
| 6 | c.3028+3A>G (p.?) | 25 | significant | splice | yes | ||
| 7 | c.3028+1delG | 71 | significant | splice | yes | ||
| 8 | c.3028G>C(p.Asp1010His) | 76 | significant | missense/splice | yes | ||
| 9 | c.3028G>T (p.Asp1010Tyr) | 71 | significant | missense/splice | yes | ||
| 10 | c.3028+1_3028+2delinsTC | 30 | significant | splice | yes | Sarcomatoid carcinoma, pleomorphic type with spindle cell and adenocarcinoma components | |
| 11 | c.3025_3028+3delGAAGGTA (p.?) | 66 | significant | splice | yes | Adenocarcinoma, lepidic predominant | |
| 12 | c.3320G>C(p.Cys1107Ser) | 52 | VUS | missense | no | ||
| 13 | c.3028+3_3028+9delinsTTTTTTT (p.?) | 34 | VUS | splice? | no | EGFR | Adenosquamous carcinoma |
| 14 | c.3082+1delG (p.?) | 44 | significant | splice | yes | Adenocarcinoma, acinar predominant | |
| 15 | c.3028+1G>C (p.?) | 30 | significant | splice | yes | Adenocarcinoma, micropapillary predominant (60%), with additional acinar (20%), solid (10%) and lepidic (10%) components | |
| 16 | c.3747G>T, (p.Trp1249Cys) | 21 | VUS | missense | no | ||
| 17 | c.3017_3028delCTTTTCCAGAAG (p.Thr1006_Asp1010delinsAsn) | 29 | significant | splice | yes | ||
| 18 | c.3028+1G>C (splice) | 20 | significant | splice | yes | ||
| 19 | c.3301G>A(p.Asp1101Asn) | 4 | VUS | missense | no | ||
| 20 | c.3028+3A>T | 24 | VUS | splice? | no | Adenocarcinoma, lepidic predominant | |
| 21 | c.3002_3027delTAGACTACCGAGCTACTTTTCCAGAA (p.Val1001Glyfs*5) | 7 | VUS | splice? | no | Adenocarcinoma (micropapillary 60%, acinar 40%, papillary 10%) | |
| 22 | c.3028G>C(p.Asp1010His) | 92 | significant | missense/splice | yes | ||
| 23 | c.3017_3028del (p.Thr1006_Asp1010delinsAsn) | 14 | VUS | splice? | no | Primary lung adenosquamous carcinoma | |
| 24 | c.3752C>T (p.Ala1251Val) | 5 | VUS | missense | no | KRAS | |
| 25 | c.3028 + 3A>G | 23 | VUS | splice? | no | Adenocarcinoma, acinar predominant (80%) with lepidic (20%) pattern | |
| 26 | c.3028G>T(p.Asp1010Tyr) | 23 | significant | missense/splice | yes | Adenocarcinoma, acinar-predominant | |
| 27 | c.3028+1G>A (p.?) | 3 | significant | splice | yes | ||
| 28 | c.3017_3028+2del | 40 | significant | splice | yes | ||
| 29 | c.3028+2T>C (p.?) | 11 | significant | splice | yes | Adenocarcinoma, solid predominant (80%) with additional acinar pattern (20%) pattern. | |
| 30 | c.3028G>A (p.Asp1010Asn) | 15 | significant | missense/splice | yes | ||
| 31 | c.3028G>C (p.Asp1010His) | 77 | significant | missense/splice | yes | ||
| 32 | c.3028+1G>C | 73 | significant | splice | yes | Adenocarcinoma, acinar predominant (60%), with additional solid (30%) and micropapillary (10%) patterns. | |
| 33 | c.3028+2T>C | 54 | significant | splice | yes | ||
| 34 | c.3028+1G>T | 25 | significant | splice | yes | Adenocarcinoma with predominant acinar pattern | |
| 35 | c.3028+2T>C (p.?) | 31 | significant | splice | yes | Adenocarcinoma, lepidic predominant (55%) with acinar pattern (45%). | |
| 36 | c.3028G>C (p.Asp1010His) | 52 | significant | missense/splice | yes | Adenocarcinoma, lepidic-predominant (70%) with acinar (20%) and solid (10%) patterns. | |
| 37 | c.3028+1G>A | 35 | significant | splice | yes | ||
| 38 | c.3028G>C (p.Asp1010His) | 3 | significant | missense/splice | yes | Adenocarcinoma, lepidic predominant (80%), with additional acinar pattern (20%). | |
| 39 | c.3027_3028+6delAAGGTATAT | 3 | significant | splice | yes | ||
| 40 | c.3281A>G (p.His1094Arg) and c.3340+1G>A (intron 16) | 4 | significant and VUS | missense | no and no | KRAS | |
| 41 | c.3007T>C(p.Tyr1003His) | 6 | significant | missense | no | ||
| 42 | c.3028G>T(p.Asp1010Tyr) | 73 | significant | missense/splice | yes | ||
| 43 | c.3028G>C | 40 | significant | splice | yes | Adenocarcinoma, lepidic predominant (60%), with additional acinar pattern (40%). | |
| 44 | c.3028+2T>C | 20 | significant | splice | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, L.; Mehrotra, H.; He, X.; Bosler, D. MET Exon 14 Variants in Non-Small Cell Lung Carcinoma: Prevalence, Clinicopathologic and Molecular Features. J. Mol. Pathol. 2023, 4, 46-56. https://doi.org/10.3390/jmp4010006
Yuan L, Mehrotra H, He X, Bosler D. MET Exon 14 Variants in Non-Small Cell Lung Carcinoma: Prevalence, Clinicopathologic and Molecular Features. Journal of Molecular Pathology. 2023; 4(1):46-56. https://doi.org/10.3390/jmp4010006
Chicago/Turabian StyleYuan, Lisi, Harshita Mehrotra, Xin He, and David Bosler. 2023. "MET Exon 14 Variants in Non-Small Cell Lung Carcinoma: Prevalence, Clinicopathologic and Molecular Features" Journal of Molecular Pathology 4, no. 1: 46-56. https://doi.org/10.3390/jmp4010006
APA StyleYuan, L., Mehrotra, H., He, X., & Bosler, D. (2023). MET Exon 14 Variants in Non-Small Cell Lung Carcinoma: Prevalence, Clinicopathologic and Molecular Features. Journal of Molecular Pathology, 4(1), 46-56. https://doi.org/10.3390/jmp4010006





