Abstract
Somatic MET exon 14 skipping mutations (MET ex14) are targetable driver mutations for non-small cell lung cancer (NSCLC), responsive to MET inhibitors. Objective: This study seeks to further characterize the clinicopathologic features and mutational profile of MET ex14 variant NSCLC. Design: Retrospective review of all MET ex14 tested NSCLC. Testing for selected BRAF, EGFR, HER2, KRAS, and MET mutations was performed using a clinically validated NGS assay, followed by MiSeq sequencing. Variants were classified as significant (Tier1/2) or variants of uncertain significance (VUS) per 2017 AMP/ASCO/CAP Joint Consensus Guidelines. PD-L1 expression was assessed by immunohistochemistry. Results: Of 2296 NSCLCs tested between 2017-7/2019, MET ex14 variants were present in 44 (1.9%). A total of 32 of 44 variants were MET exon 14 skipping, while the other 12 mutations were significant missense (3) or VUS (9). Of nine VUS, five were adjacent to the canonical splice site and likely to impact splicing. Four cases had concomitant mutations. Of 35 cases with known clinical staging, stage 1–2 = 20 (57%), stage 3 = 3 (9%), and stage 4 = 12 (34%). Of 19 resected NSCLSs, histological types and growth pattern included 7 lepidic pattern-predominant. A high percentage of tumors with MET ex14 mutations are positive for PD-L1, and the percentage of cases with PD-L1 expression >50% trends higher in more advanced disease. Conclusions: Most MET variants identified in our cohort (73%) are MET ex14 skipping. The prevalence of MET ex14 variants is 1.9%, and a large percentage of tumors has lower clinical stage and less aggressive pathologic features.
1. Introduction
The mesenchymal-to-epithelial transition (MET) proto-oncogene encodes a tyrosine kinase receptor that controls cell growth and migration and plays an important role in tumor invasion and metastasis [1]. MET exon 14 encodes the juxta membrane region of the receptor which is known to modulate receptor downregulation via Cbl binding and ubiquitin-mediated degradation; mutations to MET exon 14 exert their effect through altered splicing that results in exon 14 skipping [2]. MET exon 14 skipping leads to loss of a phosphorylation site required for Cbl binding, reducing ubiquitin-mediated degradation and promoting oncogenesis [3]. Splice site mutations in exon 14 of MET are reported to occur in 3% of lung adenocarcinomas and 7.7–32% of sarcomatoid carcinomas [4,5,6]. MET amplification can also lead to MET activation in tumors and has been reported to occur up to 5.6% of non-small cell lung cancer (NSCLC) [6,7]. Similar to other driver oncogenes such as epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK), that can be targeted with small molecule tyrosine kinase inhibitors (TKIs); driver alterations in the MET gene have also been identified as a target in NSCLC. Currently, FDA has approved Capmatinib and Tepotinib for use in patients with NSCLC harboring MET exon 14 skipping mutations [8].
With the advent of improved sequencing technologies, routine detection of MET exon 14 mutations has become more feasible. This study seeks to further characterize the clinicopathologic features and mutational profile of cases with MET ex14 variants in patients with NSCLC. As the development of acquired resistance to targeted therapy is very common in NSCLC patients with actionable driver alterations, immunotherapy also plays an important role in the management of these patients. We thus further analyzed the percentage of tumors with MET ex14 mutations that are positive for program death ligand 1 (PD-L1), and the percentage of cases with PD-L1 expression >50% in more advanced disease.
2. Subjects and Methods
A retrospective review of all NSCLC cases at the Cleveland Clinic tested for MET ex14 skipping mutations was conducted. For MET exon 14 testing, genomic DNA was extracted from FFPE or FNA tumor specimens and tested for selected BRAF, EGFR, HER2, KRAS, and MET mutations using a clinically validated targeted next generation sequencing (NGS) assay. Testing utilized the customized Cancer Hotspot Panel v1 (cCHPv1, Thermo Fisher, Waltham, MA, USA) amplification-based library preparation customized to include MET exon 14 splice site variants, followed by sequencing on the MiSeq NGS platform (Illumina, San Diego, CA, USA). A primary bioinformatics analysis was performed using MiSeq Real Time Analysis (RTA), and sequence alignment and variant calls were performed using NextGENe® software (Softgenetics®, State College, PA, USA). Coverage of MET included the following hot spots and immediately flanking sequences: codons 168, 375, 830, 991, 992, 1001–1010, 1094, 1099, 1100, 1106, 1112, 1230, 1235, 1253 and intronic loci at the intron 14 consensus donor splice site c.3028+1 and c.3028+2 (NM_000245.3). Variants were classified as significant (Tier1/2) or variants of uncertain significance (VUS) per 2017 AMP/ASCO/CAP Joint Consensus Guidelines10. ALK, RET, and ROS1 fusions were detected in parallel by FISH.
Definitions of variant categories are as follows: (1) “splice”: involve consensus splice site (−1, −2, +1, +2) or has been previously reported in the literature as exon 14 skipping; (2) “VUS, possible splice”: occurs near the exon/intron junction (in the region of other known exon 14 skipping variants) but does not involve consensus splice site (−1, −2, +1, +2) and has not been previously reported in the literature as MET exon 14 skipping; (3) “missense, significant”: has been previously reported as a significant somatic mutation in lung carcinoma but does not involve MET exon 14 splicing; (4) “VUS, missense”: not expected to involve MET exon 14 splicing and has not been previously reported as a significant somatic mutation in NSCLC. PD-L1 expression was assessed by immunohistochemistry (IHC) using a mouse monoclonal PD-L1 antibody (22-c3, Dako, CA, USA). Clinicopathologic features for MET ex14 positive cases were evaluated by chart review.
The study was performed in accordance with the Institutional review board (IRB) of the Cleveland Clinic (IRB# 19-1245).
3. Results
Of the 2296 cases of NSCLC analyzed by DNA-based NGS between 2017-7/2019, MET ex14 variants were present in 44 cases (1.9%), of which 26 (59%) were males and 18 (41%) were females. The median age of positive cases was 76 years (±9.6, 59% men; 41% women). Specimen type was FNA in 46.7%. A total of 32 of 44 variants were MET exon 14 skipping mutations (previously reported and/or involve the canonical recognition site), while the other 12 mutations were significant missense (3) or VUS (9). Of nine VUS, five were adjacent to the canonical splice site and likely to impact splicing, while four were missense variants (Figure 1A).
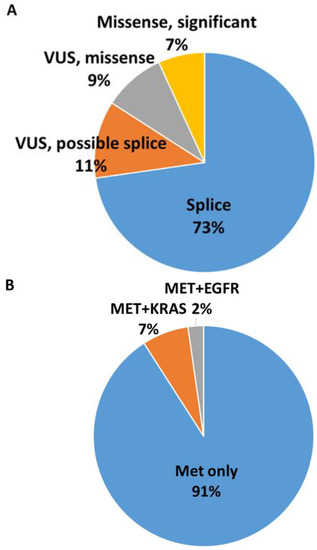
Figure 1.
(A) MET ex14 variants present in 44 cases: 32 MET exon 14 skipping, 3 significant missense, 9 VUS. Of 9 VUS, 5 were adjacent to the canonical splice site and likely to impact splicing, and 4 were missense variants. (B) Four cases had concomitant mutations including 3 with KRAS concomitant mutations and 1 with EGFR concomitant mutations; none were classic exon 14 skipping.
The average variant allele fraction (VAF) was 33.6%. Four cases (9%) had concomitant mutations (3 = KRAS, 1 = EGFR) (Figure 1B). In the three tumors with KRAS concomitant mutations, MET mutations were present at a much lower VAF compared with KRAS. However, the tumor with EGFR concomitant mutation had EGFR mutation at only 3% allele frequency but MET mutation at 34% allele frequency (supplementary information). All cases tested for ROS, RET, and ALK by FISH were negative (20 of 44 were tested for ROS, RET, and ALK, 19 were tested for ALK only, 1 was tested for RET only, and 1 was tested for RET and ROS).
Out of the 35 cases with known clinical staging, 20 cases (57%) were stage 1 or 2, 3 cases (9%) were stage 3, while 12 cases (34%) were stage 4. Of the 19 tumors that were resected, the histological types and growth pattern of NSCLC included 7 lepidic pattern-predominant, 6 acinar pattern-predominant, 2 micropapillary-predominant, 1 solid-predominant, 1 sarcomatoid, and 2 adenosquamous (Figure 2, representative cases with MET variants. Specifically, lepidic pattern, case #1; acinar pattern, case #3; micropapillary pattern, case #15; solid, case #29; sarcomatoid, case #10; and adenosquamous, case #13). PD-L1 expression was grouped into three categories, with 19% showing no expression, 41% each showing 1–49% expression and >50% expression. High PD-L1 expressing tumors showed disproportionately high tumor stage, with 4 of 6 (67%) evaluable stage 4 tumors showing >50% expression compared with 6 of 18 (33%) stage 1–3 tumors. In contrast, although four of five lepidic-predominant tumors were positive for PD-L1, they were all at the level of 1–49%. The clinicopathologic correlation is summarized in Table 1. Additional details on MET variants and clinicopathologic features are shown in Table 2.
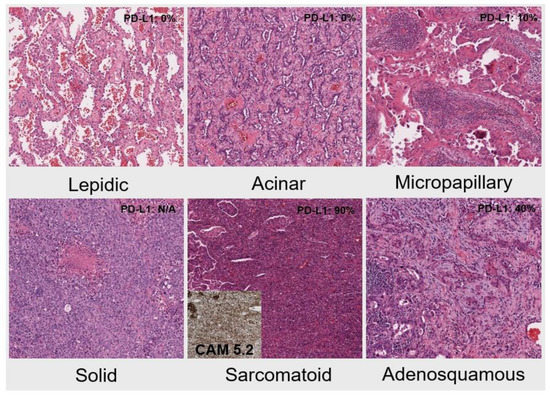
Figure 2.
The histological types and growth pattern of NSCLC include lepidic pattern-predominant, acinar pattern-predominant, micropapillary-predominant, solid-predominant, sarcomatoid (inlet: CAM5.2 immunostain), and adenosquamous (IHC, ×100).

Table 1.
Clinicopathologic correlation of tumors with METex14 mutations.

Table 2.
Additional details on MET variants and clinicopathologic features.
4. Discussion
Lung cancer continues to be the leading cause of cancer-related mortality among men and women worldwide [9], driving the need to discover new targetable genomic mutations. MET exon 14 skipping is a primary oncogenic driver sensitive to MET inhibition. In addition to existing FDA approved therapeutic options for MET exon 14 skipping positive tumors such as capmatinib, crizotinib, and tepotinib [1], several selective MET inhibitors are being investigated, including emibetuzumab and savolitinib, and are promising therapies with maximized efficacy and reduced off-target toxicity [10].
Previous studies have reported the prevalence of MET ex14 to be approximately 3.0% in NSCLC with highest prevalence in the pulmonary sarcomatoid subtype [3]. While Awad et al. [4] and Schrock et al. [5] reported a prevalence of 3% and 2.7% of MET ex14 variants, in studies conducted by Zheng et al. [11] and Liu et al. [12], the prevalence was observed to be 1.3% and 1%, respectively, possibly reflecting a lower incidence in Asians. In our cohort, MET ex14 variants were observed in 1.9% cases (44/2296). Our lower observed prevalence may be due in part to universal testing of NSCLC at our institution rather than testing of only advanced disease. Additionally, although our MET test is expected to detect most relevant exon 14 skipping variants, our lower reported prevalence may be partially attributable to variants lying outside the test’s covered regions and/or alterations not efficiently detected by next generation sequencing methodology such as large deletions.
Recent studies by Poirot et al. and Davies et al. have demonstrated the superiority of RNA-based assays in detecting MET exon 14 skipping mutations compared with DNA-based assays [13,14]. In both these studies, DNA-based assays were able to identify only approximately 60% of the mutations as any variant outside the amplified area or preventing binding due to mutation of the primer would not be detected. In contrast, in RNA-based assays, any variant causing skipping of exon 14 in vivo is identified as fusion on exon 13 to exon 15. In general, RNA-based targeted approach analyses and quantifies directly fusion transcripts and is more accurate than DNA panels on tumor tissue, but it can be limited by RNA quality and quantity. However, amplicon-based RNA NGS panels can detect gene fusions in poor quality RNA samples, such as those obtained from FFPE tissue samples (PMID: 32726941). This is a major limitation of our study, as adding RNA-based NGS assays or using combined DNA-RNA testing might have allowed us to better classify the samples with non-canonical DNA mutations, including missense and VUS variants in this cohort.
In contrast with other targetable mutations seen in NSCLC such as ALK and ROS1 rearrangements and KRAS, EGFR, and BRAF mutations, MET ex14 mutations have been identified in older patients. In a review of 933 patients with NSCLC, Awad et al. identified MET exon 14 skipping mutations in 28 patients with a median age of 72.5 years [4]. Whereas 68% and 60.4% patients with MET ex14 variants were females in the study conducted by Awad et al. [4] and Shrock et al. [5], respectively, in a study conducted by Wang et al., 66% patients were males [15]. In our cohort, the mean age of the patients was 76 years (53 to 91 years) which is consistent with previous studies. However, there is male preference is our study, with 59% (26) males and 41% (18) females.
MET ex14 mutations have been observed across various histological subtypes of non-small cell lung carcinoma, with a decreasing order of prevalence seen in sarcomatoid carcinoma, adenosquamous carcinoma, adenocarcinoma, and squamous cell carcinoma [4,16]. In our study, of the 19 patients who underwent resection, 16 patients had adenocarcinoma (including five tumors with lepidic-predominant pattern), 2 adenosquamous carcinoma, and 1 sarcomatoid carcinoma. As primary lung tumors, adenosquamous carcinoma and sarcomatoid carcinoma are relatively rare, comprising 0.3–1.3% and 0.4–4% of all lung carcinomas, respectively [17,18]. Our data are consistent with the previous studies showing that MET ex14 appears enriched in these two aggressive histologic subtypes [17,18]. However, a large percentage of tumors also had lower clinical stage and less aggressive pathologic features, both possibly reflecting sampling differences attributed to universal testing of NSCLC at our institution rather than testing of only advanced diseases was common in early studies due to the emphasis on targeted therapies for advanced disease. This seemingly paradoxical observation is consistent with studies from other centers where MET ex14 variants testing is performed routinely for all cases of NSCLC [4].
The spectrum of known MET exon 14 skipping mutations must be considered when designing diagnostic tests for their detection. In literature, hundreds of distinct genetic alterations leading to MET exon 14 skipping have been reported, including base substitutions and insertions or deletions at the splice acceptor site, at the splice donor site, and in intronic non-coding regions immediately adjacent to the splice acceptor site, as well as whole exon deletions [19]. In our cohort, 32 of the 44 (73%) MET ex14 mutations were previously reported and/or canonical recognition sites skipping variants. Nine patients had VUS, five of which were adjacent to a canonical splice site and were likely to impact splicing and four were missense variants. Three patients had significant missense mutations. To detect these potential alterations, it is important to sequence both exon 14 and its surrounding regions, although a plurality of MET exon 14 skipping events occur at the donor site [20].
MET exon 14 skipping mutations are usually mutually exclusive of other characteristic driver mutations in NSCLC. Whereas the cancer genome atlas research network [21] and Awad et al. [4] identified no concomitant mutation with MET ex14 skipping mutation, in the study conducted by Shrock et al., concurrent KRAS mutation and EGFR amplification was observed in 3% and 6.4% cases, respectively [5]. In our cohort, four cases (9%) had concomitant mutations with three harboring KRAS mutations and one with EGFR mutation. However, none of the co-mutations occurred with a known functional pathogenic MET ex14 skipping mutation. The EGFR mutation was co-mutated with a VUS possible splice of MET ex14. The KRAS mutations were co-mutated with a significant missense mutation of METe x14, a VUS missense of MET ex14, and a VUS missense mutation together with a possible splice site mutation of MET exon 16, respectively. Further investigations of these cases would be required to confidently establish true functionally co-mutated status. Moreover, the testing methodology used does not distinguish whether the mutations are part of the same clone (co-mutational status as a subclone) or reflect tumoral heterogeneity.
The development of monoclonal antibodies against PD-1 receptor and its ligand, PD-L1 has provided a major breakthrough in the management of patients with NSCLC in the last decade. In a review of 147 patients with MET exon 14 skipping mutations, PD-L1 expression of > 1% was seen in 57 0f 111 patients, although the overall survival did not improve in patients who were administered PD-L1 immunotherapy [22]. In a similar study conducted by Xu et al., 205 of 401 patients with NSCLC and MET exon14 skipping had PD-L1 expression of > 1% [23]. In our cohort, 27 patients had tissue available for evaluation of PD-L1 expression. PD-L1 expression of 0%, 1–49%, and >50% was observed in 19% (5/27), 41% (11/27), and 41% (11/27) of patients, respectively, which is higher compared with previous studies (approximately 80% versus 50%). Patients with stage 4 tumors showed a significantly higher percentage of PD-L1 expression of >50% (67%, 4/6) compared with stage 1–3 tumors (33%, 6/18). In contrast, although four of five lepidic-predominant tumors were positive for PD-L1, they were all at the level of 1–49%.
Engagement of programmed cell death-1 (PD-1) receptor by its ligands PD-L1 is an important adaptor immune mechanism by tumor cells. This leads to downregulation of T-cells in the tumor microenvironment which can be reversed by PD1 and PD-L1 blocking antibodies such as pembrolizumab, nivolumab, atezolizumab, and durvalumab [24]. These have shown promise in the treatment of patients with non-small cell lung carcinoma with response rates of over 20% in treatment-naïve patients [25]. Testing for PD-L1 expression remains current standard in patients with NSCLC who are likely to respond to immunotherapy. It has been seen that in patients with higher PD-L1 expression have better response to immunotherapy than patients with lower PD-L1 expression [26,27]. Presently, IHC remains the gold standard for quantifying PD-L1 expression in tumor samples [28]. Although these findings have potential therapeutic implications, NCCN recognizes that “Patients with MET ex14 skipping mutations have a modest response (16%, single-agent ICIs) to immunotherapy, even those with high PD-L1 levels.” (NCCN NSCLC version 1.2023, MS-21). Additionally, the guidelines support that any patient with a targetable oncogenic driver should receive targeted therapy before immunotherapy. Larger studies are required to clarify the PD-L1 status in patients with MET exon 14 skipping mutations and the potential contribution of immunotherapies in these patients, especially in patients with high-stage diseases.
In conclusion, roughly three quarters of MET variants identified in our cohort are MET ex14 skipping, another one tenth likely result in exon 14 skipping, while the other 16% are missense variants presumably unrelated to splicing. Our prevalence of MET ex14 variants is 1.9%, and a large percentage of tumors has lower clinical stage and less aggressive pathologic features. Notably, a high percentage of tumors with MET ex14 mutations are positive for PD-L1, and the percentage of cases with PD-L1 expression >50% trends higher in more advanced stage disease. It is unclear whether certain MET exon 14 mutations are more responsive to c-Met inhibition than others and more prospective clinical trials will be necessary to determine if immunotherapy or combination strategies (i.e., immunotherapy and chemotherapy) in addition to targeted therapy in this population will provide survival advantage. The presence of existing and potential targeted therapies for MET inhibition highlight the importance of including testing for MET in the molecular evaluation of NSCLC.
Author Contributions
L.Y. and D.B. contributed to study conception, data acquisition, data analysis and interpretation, and manuscript writing and editing. H.M. contributed to manuscript writing and editing. X.H. contributed to data acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was performed in accordance with the Institutional review board (IRB) of the Cleveland Clinic (IRB# 19-1245).
Informed Consent Statement
No patient involved.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fujino, T.; Suda, K.; Mitsudomi, T. Lung Cancer with MET exon 14 Skipping Mutation: Genetic Feature, Current Treatments, and Future Challenges. Lung Cancer: Targets Ther. 2021, 12, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zou, Q.; Liu, H.; Qiu, B.; Li, Q.; Lin, Y.; Liang, Y. Management of Non-small Cell Lung Cancer Patients with MET Exon 14 Skipping Mutations. Curr. Treat. Options Oncol. 2020, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Ho, A.T.N.; Altibi, A.M.; Nakazawa, T.; Katoh, R.; Kondo, T. Clinicopathological implications of MET exon 14 mutations in non-small cell lung cancer—A systematic review and meta-analysis. Lung Cancer 2018, 123, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET Exon 14 Mutations in Non–Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef]
- Schrock, A.B.; Frampton, G.M.; Suh, J.; Chalmers, Z.R.; Rosenzweig, M.; Erlich, R.L.; Halmos, B.; Goldman, J.; Forde, P.; Leuenberger, K.; et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J. Thorac. Oncol. 2016, 11, 1493–1502. [Google Scholar] [CrossRef]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.H.; Chung, L.Y.; Chau, S.L.; Lung, R.W.M.; Tong, C.Y.; Chow, C.; Tin, E.K.Y.; Yu, Y.H.; et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non–Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef]
- Ou, S.-H.I.; Kwak, E.L.; Siwak-Tapp, C.; Dy, J.; Bergethon, K.; Clark, J.W.; Camidge, D.R.; Solomon, B.J.; Maki, R.G.; Bang, Y.-J.; et al. Activity of Crizotinib (PF02341066), a Dual Mesenchymal-Epithelial Transition (MET) and Anaplastic Lymphoma Kinase (ALK) Inhibitor, in a Non-small Cell Lung Cancer Patient with De Novo MET Amplification. J. Thorac. Oncol. 2011, 6, 942–946. [Google Scholar] [CrossRef]
- Jørgensen, J.T.; Mollerup, J. Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC. Cancers 2022, 14, 2150. [Google Scholar] [CrossRef]
- Lung Cancer Statistics. Available online: https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html (accessed on 14 February 2021).
- Salgia, R.; Sattler, M.; Scheele, J.; Stroh, C.; Felip, E. The promise of selective MET inhibitors in non-small cell lung cancer with MET exon 14 skipping. Cancer Treat. Rev. 2020, 87, 102022. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, R.; Ye, T.; Yu, S.; Hu, H.; Shen, X.; Li, Y.; Ji, H.; Sun, Y.; Chen, H. MET exon 14 skipping defines a unique molecular class of non-small cell lung cancer. Oncotarget 2016, 7, 41691–41702. [Google Scholar] [CrossRef]
- Liu, S.-Y.; Gou, L.-Y.; Li, A.-N.; Lou, N.-N.; Gao, H.-F.; Su, J.; Yang, J.-J.; Zhang, X.-C.; Shao, Y.; Dong, Z.-Y.; et al. The Unique Characteristics of MET Exon 14 Mutation in Chinese Patients with NSCLC. J. Thorac. Oncol. 2016, 11, 1503–1510. [Google Scholar] [CrossRef]
- Poirot, B.; Doucet, L.; Benhenda, S.; Champ, J.; Meignin, V.; Lehmann-Che, J. MET Exon 14 Alterations and New Resistance Mutations to Tyrosine Kinase Inhibitors: Risk of Inadequate Detection with Current Amplicon-Based NGS Panels. J. Thorac. Oncol. 2017, 12, 1582–1587. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.D.; Lomboy, A.; Lawrence, C.A.; Yourshaw, M.; Bocsi, G.T.; Camidge, D.R.; Aisner, D.L. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. J. Thorac. Oncol. 2019, 14, 737–741. [Google Scholar] [CrossRef]
- Wang, S.X.; Zhang, B.M.; Wakelee, H.A.; Koontz, M.Z.; Pan, M.; Diehn, M.; Kunder, C.A.; Neal, J.W. Case series of MET exon 14 skipping mutation-positive non-small-cell lung cancers with response to crizotinib and cabozantinib. Anti-Cancer Drugs 2019, 30, 537–541. [Google Scholar] [CrossRef]
- Saffroy, R.; Fallet, V.; Girard, N.; Mazieres, J.; Sibilot, D.M.; Lantuejoul, S.; Rouquette, I.; Thivolet-Bejui, F.; Vieira, T.; Antoine, M.; et al. MET exon 14 mutations as targets in routine molecular analysis of primary sarcomatoid carcinoma of the lung. Oncotarget 2017, 8, 42428–42437. [Google Scholar] [CrossRef] [PubMed]
- Filosso, P.L.; Ruffini, E.; Asioli, S.; Giobbe, R.; Macri, L.; Bruna, M.C.; Sandri, A.; Oliaro, A. Adenosquamous lung carcinomas: A histologic subtype with poor prognosis. Lung Cancer 2011, 74, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Maneenil, K.; Xue, Z.; Liu, M.; Boland, J.; Wu, F.; Stoddard, S.M.; Molina, J.; Yang, P. Sarcomatoid Carcinoma of the Lung: The Mayo Clinic Experience in 127 Patients. Clin. Lung Cancer 2018, 19, e323–e333. [Google Scholar] [CrossRef]
- Socinski, M.A.; Pennell, N.A.; Davies, K.D. METExon 14 Skipping Mutations in Non–Small-Cell Lung Cancer: An Overview of Biology, Clinical Outcomes, and Testing Considerations. JCO Precis. Oncol. 2021, 5, 653–663. [Google Scholar] [CrossRef]
- Awad, M.M.; Lee, J.K.; Madison, R.; Classon, A.; Kmak, J.; Frampton, G.M.; Alexander, B.M.; Venstrom, J.; Schrock, A.B. Characterization of 1,387 NSCLCs with MET exon 14 (METex14) skipping alterations (SA) and potential acquired resistance (AR) mechanisms. J. Clin. Oncol. 2020, 38, 9511. [Google Scholar] [CrossRef]
- Cohen, D.; Hondelink, L.M.; Solleveld-Westerink, N.; Uljee, S.M.; Ruano, D.; Cleton-Jansen, A.-M.; von der Thüsen, J.H.; Ramai, S.R.S.; Postmus, P.E.; van Roggen, J.F.G.; et al. Optimizing Mutation and Fusion Detection in NSCLC by Sequential DNA and RNA Sequencing. J. Thorac. Oncol. 2020, 15, 1000–1014. [Google Scholar] [CrossRef]
- Sabari, J.; Leonardi, G.; Shu, C.; Umeton, R.; Montecalvo, J.; Ni, A.; Chen, R.; Dienstag, J.; Mrad, C.; Bergagnini, I.; et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann. Oncol. 2018, 29, 2085–2091. [Google Scholar] [CrossRef]
- Xu, Z.; Li, H.; Dong, Y.; Cheng, P.; Luo, F.; Fu, S.; Gao, M.; Kong, L.; Che, N. Incidence and PD-L1 Expression of MET 14 Skipping in Chinese Population: A Non-Selective NSCLC Cohort Study Using RNA-Based Sequencing. OncoTargets Ther. 2020, 13, 6245–6253. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.A.; Weiss, J. Advances in the Treatment of Non–Small Cell Lung Cancer. Clin. Chest Med. 2020, 41, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Hellmann, M.D.; Rizvi, N.A.; Carcereny, E.; Leighl, N.B.; Ahn, M.-J.; Eder, J.P.; Balmanoukian, A.S.; Aggarwal, C.; Horn, L.; et al. Five-Year Overall Survival for Patients With Advanced Non-Small-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J. Clin. Oncol. 2019, 37, 2518–2527. [Google Scholar] [CrossRef] [PubMed]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Kurata, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018, 379, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Imyanitov, E.N.; Iyevleva, A.G.; Levchenko, E.V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol. 2021, 157, 103194. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).