Next Generation Digital Pathology: Emerging Trends and Measurement Challenges for Molecular Pathology
Abstract
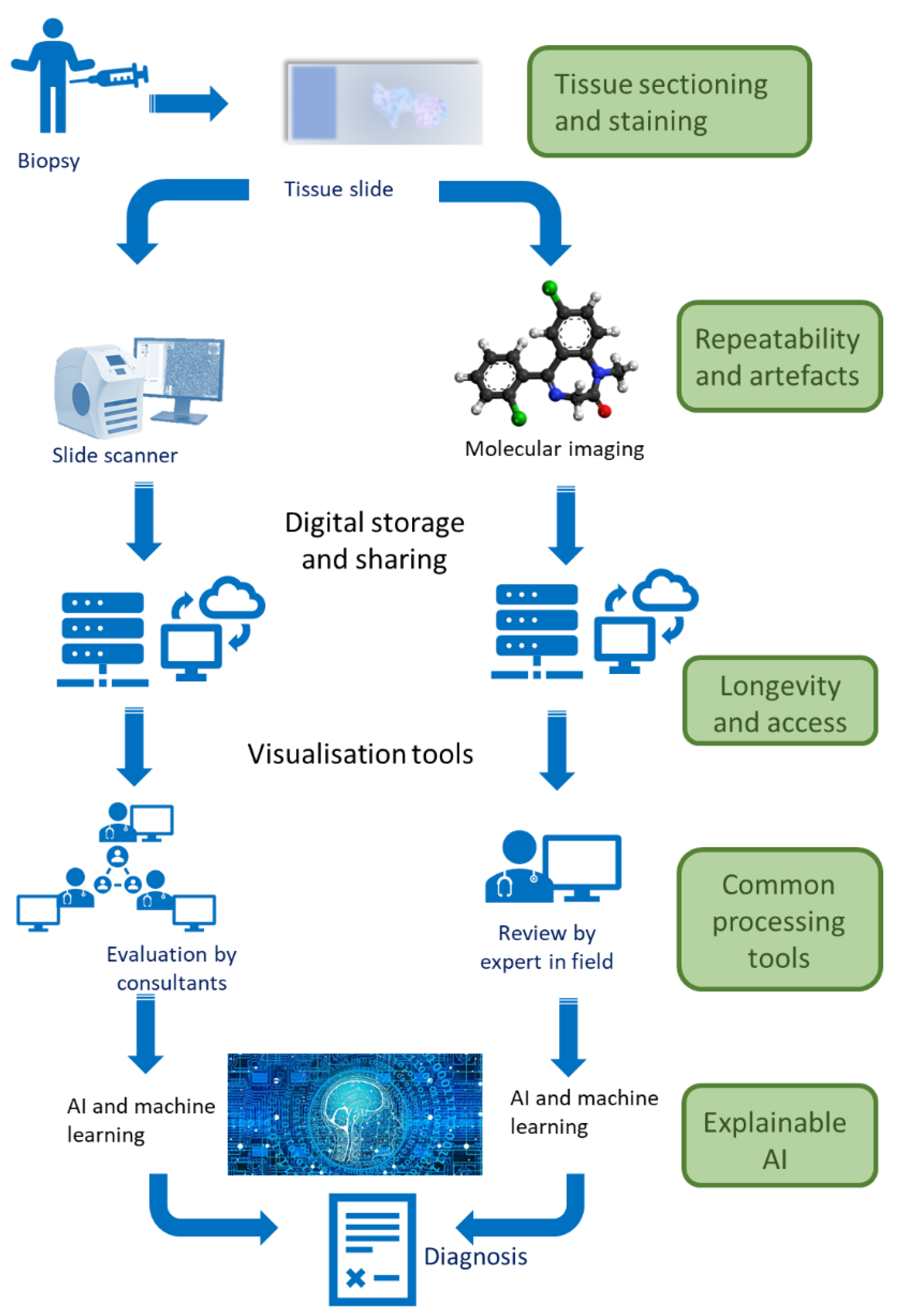
:1. What Are Digital and Molecular Pathology?
2. Why Is It Important?
3. Classical Digital Pathology
3.1. Slide Scanning
3.2. Guided Visualisation
3.3. AI for Classification
3.4. Metadata
4. Molecular Pathology
4.1. Spatial Resolution
4.2. Data Analysis
4.2.1. Pre-Processing
4.2.2. Feature Selection
4.2.3. Segmentation
4.2.4. Classification
4.2.5. Validation and Review
4.2.6. Metadata in Molecular Pathology
4.3. Barriers to Molecular Pathology
4.4. Integration of Molecular Pathology with Classical Histopathology
4.5. Routes to Standards in Molecular Pathology
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farahani, N.; Parwani, A.V.; Pantanowitz, L. Whole slide imaging in pathology: Advantages, limitations, and emerging perspectives. Pathol. Lab. Med. Int. 2015, 7, 23–33. [Google Scholar]
- Herrmann, M.D.; Clunie, D.A.; Fedorov, A.; Doyle, S.W.; Pieper, S.; Klepeis, V.; Le, L.P.; Mutter, G.L.; Milstone, D.S.; Schultz, T.J. Implementing the DICOM standard for digital pathology. J. Pathol. Inform. 2018, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.J.; Salas, L.A.; Christensen, B.C.; Sriharan, A.; Vaickus, L.J. PathFlowAI: A high-throughput workflow for preprocessing, deep learning and interpretation in digital pathology. In Proceedings of the Pacific Symposium on Biocomputing 2020, Kohala Coast, HI, USA, 3–7 January 2020; pp. 403–414. [Google Scholar]
- Yagi, Y. Color standardization and optimization in whole slide imaging. In Diagnostic Pathology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–12. [Google Scholar]
- AlZubaidi, A.K.; Sideseq, F.B.; Faeq, A.; Basil, M. Computer aided diagnosis in digital pathology application: Review and perspective approach in lung cancer classification. In Proceedings of the 2017 Annual Conference on New Trends In Information & Communications Technology Applications (NTICT), Baghdad, Iraq, 7–9 March 2017; pp. 219–224. [Google Scholar]
- Abdelmoula, W.M.; Balluff, B.; Englert, S.; Dijkstra, J.; Reinders, M.J.; Walch, A.; McDonnell, L.A.; Lelieveldt, B.P. Data-driven identification of prognostic tumor subpopulations using spatially mapped t-SNE of mass spectrometry imaging data. Proc. Natl. Acad. Sci. USA 2016, 113, 12244–12249. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, R.; Madabhushi, A. Emerging themes in image informatics and molecular analysis for digital pathology. Annu. Rev. Biomed. Eng. 2016, 18, 387–412. [Google Scholar] [CrossRef]
- Gu, J.; Taylor, C.R. Practicing pathology in the era of big data and personalized medicine. Appl. Immunohistochem. Mol. Morphol. AIMM/Off. Publ. Soc. Appl. Immunohistochem. 2014, 22, 1. [Google Scholar] [CrossRef]
- Baidoshvili, A.; Bucur, A.; van Leeuwen, J.; van der Laak, J.; Kluin, P.; van Diest, P.J. Evaluating the benefits of digital pathology implementation: Time savings in laboratory logistics. Histopathology 2018, 73, 784–794. [Google Scholar] [CrossRef]
- Vodovnik, A. Diagnostic time in digital pathology: A comparative study on 400 cases. J. Pathol. Inform. 2016, 7, 4. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Hartman, D.; Qi, Y.; Cho, E.Y.; Suh, B.; Paeng, K.; Dhir, R.; Michelow, P.; Hazelhurst, S.; Song, S.Y. Accuracy and efficiency of an artificial intelligence tool when counting breast mitoses. Diagn. Pathol. 2020, 15, 80. [Google Scholar] [CrossRef]
- Perincheri, S.; Levi, A.W.; Celli, R.; Gershkovich, P.; Rimm, D.; Morrow, J.S.; Rothrock, B.; Raciti, P.; Klimstra, D.; Sinard, J. An independent assessment of an artificial intelligence system for prostate cancer detection shows strong diagnostic accuracy. Mod. Pathol. 2021, 34, 1588–1595. [Google Scholar] [CrossRef]
- Fontelo, P.; Liu, F.; Yagi, Y. Evaluation of a smartphone for telepathology: Lessons learned. J. Pathol. Inform. 2015, 6, 35. [Google Scholar] [CrossRef]
- Grote, A.; Schaadt, N.S.; Forestier, G.; Wemmert, C.; Feuerhake, F. Crowdsourcing of histological image labeling and object delineation by medical students. IEEE Trans. Med. Imaging 2018, 38, 1284–1294. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, P.W.; Wang, Y.; McCullough, S.J. Virtual microscopy and digital pathology in training and education. Apmis 2012, 120, 305–315. [Google Scholar] [CrossRef]
- Lu, F.-K.; Calligaris, D.; Olubiyi, O.I.; Norton, I.; Yang, W.; Santagata, S.; Xie, X.S.; Golby, A.J.; Agar, N.Y. Label-free neurosurgical pathology with stimulated Raman imaging. Cancer Res. 2016, 76, 3451–3462. [Google Scholar] [CrossRef]
- Evans, A.J.; Salama, M.E.; Henricks, W.H.; Pantanowitz, L. Implementation of whole slide imaging for clinical purposes: Issues to consider from the perspective of early adopters. Arch. Pathol. Lab. Med. 2017, 141, 944–959. [Google Scholar] [CrossRef]
- Rhoads, D.D.; Mathison, B.A.; Bishop, H.S.; da Silva, A.J.; Pantanowitz, L. Review of telemicrobiology. Arch. Pathol. Lab. Med. 2016, 140, 362–370. [Google Scholar] [CrossRef]
- Janowczyk, A.; Zuo, R.; Gilmore, H.; Feldman, M.; Madabhushi, A. HistoQC: An open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inform. 2019, 3, 1–7. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Deroulers, C.; Ameisen, D.; Badoual, M.; Gerin, C.; Granier, A.; Lartaud, M. Analyzing huge pathology images with open source software. Diagn. Pathol. 2013, 8, 92. [Google Scholar] [CrossRef]
- Sakamoto, T.; Furukawa, T.; Lami, K.; Pham, H.H.N.; Uegami, W.; Kuroda, K.; Kawai, M.; Sakanashi, H.; Cooper, L.A.D.; Bychkov, A. A narrative review of digital pathology and artificial intelligence: Focusing on lung cancer. Transl. Lung Cancer Res. 2020, 9, 2255–2276. [Google Scholar] [CrossRef]
- Marti-Aguado, D.; Rodríguez-Ortega, A.; Mestre-Alagarda, C.; Bauza, M.; Valero-Pérez, E.; Alfaro-Cervello, C.; Benlloch, S.; Pérez-Rojas, J.; Ferrández, A.; Alemany-Monraval, P. Digital pathology: Accurate technique for quantitative assessment of histological features in metabolic-associated fatty liver disease. Aliment. Pharmacol. Ther. 2021, 53, 160–171. [Google Scholar] [CrossRef]
- Barisoni, L.; Hodgin, J.B. Digital pathology in nephrology clinical trials, research, and pathology practice. Curr. Opin. Nephrol. Hypertens. 2017, 26, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, B.A.; Afify, H.M. Automated classification of bacterial images extracted from digital microscope via bag of words model. In Proceedings of the 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), Cairo, Egypt, 20–22 December 2018; pp. 86–89. [Google Scholar]
- Rathore, S.; Iftikhar, M.A.; Chaddad, A.; Niazi, T.; Karasic, T.; Bilello, M. Segmentation and grade prediction of colon cancer digital pathology images across multiple institutions. Cancers 2019, 11, 1700. [Google Scholar] [CrossRef] [PubMed]
- Ertosun, M.G.; Rubin, D.L. Automated grading of gliomas using deep learning in digital pathology images: A modular approach with ensemble of convolutional neural networks. AMIA Annu. Symp. Proc. 2015, 2015, 1899–1908. [Google Scholar] [PubMed]
- Abdolahi, M.; Salehi, M.; Shokatian, I.; Reiazi, R. Artificial intelligence in automatic classification of invasive ductal carcinoma breast cancer in digital pathology images. Med. J. Islamic Repub. Iran 2020, 34, 965–973. [Google Scholar] [CrossRef]
- Puchalski, R.B.; Shah, N.; Miller, J.; Dalley, R.; Nomura, S.R.; Yoon, J.-G.; Smith, K.A.; Lankerovich, M.; Bertagnolli, D.; Bickley, K. An anatomic transcriptional atlas of human glioblastoma. Science 2018, 360, 660–663. [Google Scholar] [CrossRef]
- Aubreville, M.; Bertram, C.A.; Donovan, T.A.; Marzahl, C.; Maier, A.; Klopfleisch, R. A completely annotated whole slide image dataset of canine breast cancer to aid human breast cancer research. Sci. Data 2020, 7, 417. [Google Scholar] [CrossRef]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M. Bach: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- Good, B.M.; Su, A.I. Crowdsourcing for bioinformatics. Bioinformatics 2013, 29, 1925–1933. [Google Scholar] [CrossRef]
- Sabou, M.; Bontcheva, K.; Derczynski, L.; Scharl, A. Corpus Annotation through Crowdsourcing: Towards Best Practice Guidelines. In Proceedings of the LREC, Reykjavik, Iceland, 26–31 May 2014; pp. 859–866. [Google Scholar]
- Pocevičiūtė, M.; Eilertsen, G.; Lundström, C. Survey of XAI in digital pathology. In Artificial Intelligence and Machine Learning for Digital Pathology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 56–88. [Google Scholar]
- Santos, M.; Sá-Couto, P.; Silva, A.; Rocha, N. DICOM metadata-mining in PACS for computed radiography X-ray exposure analysis: A mammography multisite study. In Proceedings of the European Congress of Radiology-ECR 2014, Vienna, Austria, 6–10 March 2014. [Google Scholar]
- Thomas, S.A. Combining Image Features and Patient Metadata to Enhance Transfer Learning. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 26 July 2021; pp. 2660–2663. [Google Scholar]
- Thomas, S.A. Enhanced Transfer Learning Through Medical Imaging and Patient Demographic Data Fusion. arXiv 2021, arXiv:2111.14388. [Google Scholar]
- Lee, M.; Herrington, C.S.; Ravindra, M.; Sepp, K.; Davies, A.; Hulme, A.N.; Brunton, V.G. Recent advances in the use of stimulated Raman scattering in histopathology. Analyst 2021, 146, 789–802. [Google Scholar] [CrossRef]
- Orringer, D.A.; Pandian, B.; Niknafs, Y.S.; Hollon, T.C.; Boyle, J.; Lewis, S.; Garrard, M.; Hervey-Jumper, S.L.; Garton, H.J.; Maher, C.O. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 2017, 1, 0027. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Suo, S.; Tam, P.P.; Han, J.-D.J.; Peng, G.; Jing, N. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017, 12, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Asp, M.; Bergenstråhle, J.; Lundeberg, J. Spatially resolved transcriptomes—Next generation tools for tissue exploration. BioEssays 2020, 42, 1900221. [Google Scholar] [CrossRef] [PubMed]
- Diem, M.; Miljković, M.; Bird, B.; Chernenko, T.; Schubert, J.; Marcsisin, E.; Mazur, A.; Kingston, E.; Zuser, E.; Papamarkakis, K. Applications of infrared and Raman microspectroscopy of cells and tissue in medical diagnostics: Present status and future promises. Spectrosc. Int. J. 2012, 27, 463–496. [Google Scholar] [CrossRef]
- Tan, W.C.C.; Nerurkar, S.N.; Cai, H.Y.; Ng, H.H.M.; Wu, D.; Wee, Y.T.F.; Lim, J.C.T.; Yeong, J.; Lim, T.K.H. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020, 40, 135–153. [Google Scholar] [CrossRef]
- Chang, Q.; Ornatsky, O.I.; Siddiqui, I.; Loboda, A.; Baranov, V.I.; Hedley, D.W. Imaging mass cytometry. Cytom. Part A 2017, 91, 160–169. [Google Scholar] [CrossRef]
- van Hove, E.R.A.; Smith, D.F.; Heeren, R.M. A concise review of mass spectrometry imaging. J. Chromatogr. A 2010, 1217, 3946–3954. [Google Scholar] [CrossRef]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A. Optical coherence tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef]
- Xu, M.; Wang, L.V. Photoacoustic imaging in biomedicine. Rev. Sci. Instrum. 2006, 77, 041101. [Google Scholar] [CrossRef]
- Junttila, M.R.; De Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef]
- Quinn, M.; Gnan, N.; James, S.; Ninarello, A.; Sciortino, F.; Zaccarelli, E.; McManus, J. How fluorescent labelling alters the solution behaviour of proteins. Phys. Chem. Chem. Phys. 2015, 17, 31177–31187. [Google Scholar] [CrossRef]
- Dexter, A.; Steven, R.T.; Patel, A.; Dailey, L.A.; Taylor, A.J.; Ball, D.; Klapwijk, J.; Forbes, B.; Page, C.P.; Bunch, J. Imaging drugs, metabolites and biomarkers in rodent lung: A DESI MS strategy for the evaluation of drug-induced lipidosis. Anal. Bioanal. Chem. 2019, 411, 8023–8032. [Google Scholar] [CrossRef] [Green Version]
- Sauer, S.; Freiwald, A.; Maier, T.; Kube, M.; Reinhardt, R.; Kostrzewa, M.; Geider, K. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS ONE 2008, 3, e2843. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, R.; Prabitha, V.G.; Dora, T.K.; Chopra, S.; Maheshwari, A.; Deodhar, K.; Rekhi, B.; Sukumar, N.; Krishna, C.M.; Subhash, N. A comparative evaluation of diffuse reflectance and Raman spectroscopy in the detection of cervical cancer. J. Biophotonics 2017, 10, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, F. The effectiveness of spectral similarity measures for the analysis of hyperspectral imagery. Int. J. Appl. Earth Obs. Geoinf. 2006, 8, 3–17. [Google Scholar] [CrossRef]
- Keogh, E.; Mueen, A. Curse of dimensionality. In Encyclopedia of Machine Learning; Springer: Berlin/Heidelberg, Germany, 2011; pp. 257–258. [Google Scholar]
- Kilkenny, M.F.; Robinson, K.M. Data quality:“Garbage in–garbage out”. Health Inf. Manag. J. 2018, 47, 103–105. [Google Scholar] [CrossRef]
- Lasch, P. Spectral pre-processing for biomedical vibrational spectroscopy and microspectroscopic imaging. Chemom. Intell. Lab. Syst. 2012, 117, 100–114. [Google Scholar] [CrossRef]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2, 1–38. [Google Scholar] [CrossRef]
- Coombes, K.R.; Baggerly, K.A.; Morris, J.S. Pre-processing mass spectrometry data. In Fundamentals of Data Mining in Genomics and Proteomics; Springer: Berlin/Heidelberg, Germany, 2007; pp. 79–102. [Google Scholar]
- Auer, N.; Hrdina, A.; Hiremath, C.; Vcelar, S.; Baumann, M.; Borth, N.; Jadhav, V. ChromaWizard: An open source image analysis software for multicolor fluorescence in situ hybridization analysis. Cytom. Part A 2018, 93, 749–754. [Google Scholar] [CrossRef]
- Saafin, W.; Schaefer, G. Pre-processing techniques for colour digital pathology image analysis. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis, Edinburgh, UK, 11–13 July 2017; pp. 551–560. [Google Scholar]
- Avila Cobos, F.; Alquicira-Hernandez, J.; Vandesompele, J.; Powell, J.; Mestdagh, P.; De Preter, K. Benchmarking the impact of data transformation, pre-processing and choice of method in the computational deconvolution of transcriptomics data. In Proceedings of the ISMB/ECCB 2019, Basel, Switzerland, 21–25 July 2019. [Google Scholar]
- Murta, T.; Steven, R.T.; Nikula, C.J.; Thomas, S.A.; Zeiger, L.B.; Dexter, A.; Elia, E.A.; Yan, B.; Campbell, A.D.; Goodwin, R.J. Implications of Peak Selection in the Interpretation of Unsupervised Mass Spectrometry Imaging Data Analyses. Anal. Chem. 2021, 93, 2309–2316. [Google Scholar] [CrossRef] [PubMed]
- Salomatina, E.V.; Jiang, B.; Novak, J.; Yaroslavsky, A.N. Optical properties of normal and cancerous human skin in the visible and near-infrared spectral range. J. Biomed. Opt. 2006, 11, 064026. [Google Scholar] [CrossRef]
- Clark, A.R.; Calligaris, D.; Regan, M.S.; Krummel, D.P.; Agar, J.N.; Kallay, L.; MacDonald, T.; Schniederjan, M.; Santagata, S.; Pomeroy, S.L. Rapid discrimination of pediatric brain tumors by mass spectrometry imaging. J. Neuro-Oncol. 2018, 140, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Van Der Maaten, L.; Postma, E.; Van den Herik, J. Dimensionality reduction: A comparative. J Mach Learn Res 2009, 10, 66–71. [Google Scholar]
- Dexter, A.; Thomas, S.A.; Steven, R.T.; Robinson, K.N.; Taylor, A.J.; Elia, E.; Nikula, C.; Campbell, A.D.; Panina, Y.; Najumudeen, A.K. Training a neural network to learn other dimensionality reduction removes data size restrictions in bioinformatics and provides a new route to exploring data representations. bioRxiv 2020. [Google Scholar] [CrossRef]
- He, W.; Zhang, H.; Zhang, L.; Philips, W.; Liao, W. Weighted sparse graph based dimensionality reduction for hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2016, 13, 686–690. [Google Scholar] [CrossRef]
- Inglese, P.; Correia, G.; Takats, Z.; Nicholson, J.K.; Glen, R.C. SPUTNIK: An R package for filtering of spatially related peaks in mass spectrometry imaging data. Bioinformatics 2019, 35, 178–180. [Google Scholar] [CrossRef]
- Kobrina, Y.; Rieppo, L.; Saarakkala, S.; Jurvelin, J.S.; Isaksson, H. Clustering of infrared spectra reveals histological zones in intact articular cartilage. Osteoarthr. Cartil. 2012, 20, 460–468. [Google Scholar] [CrossRef]
- Krafft, C.; Steiner, G.; Beleites, C.; Salzer, R. Disease recognition by infrared and Raman spectroscopy. J. Biophotonics 2009, 2, 13–28. [Google Scholar] [CrossRef]
- Maurer, C.; Holmstrom, S.R.; He, J.; Laise, P.; Su, T.; Ahmed, A.; Hibshoosh, H.; Chabot, J.A.; Oberstein, P.E.; Sepulveda, A.R. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 2019, 68, 1034–1043. [Google Scholar] [CrossRef]
- Dexter, A.; Race, A.M.; Steven, R.T.; Barnes, J.R.; Hulme, H.; Goodwin, R.J.; Styles, I.B.; Bunch, J. Two-phase and graph based clustering methods for accurate and efficient segmentation of large mass spectrometry images. Anal. Chem. 2017, 89, 11293–11300. [Google Scholar] [CrossRef]
- Dexter, A.; Race, A.; Styles, I.; Bunch, J. Testing for multivariate normality in mass spectrometry imaging data: A robust statistical approach for clustering evaluation and the generation of synthetic mass spectrometry imaging datasets. Anal. Chem. 2016, 88, 10893–10899. [Google Scholar] [CrossRef]
- Race, A.M.; Sutton, D.; Hamm, G.; Maglennon, G.; Morton, J.P.; Strittmatter, N.; Campbell, A.; Sansom, O.J.; Wang, Y.; Barry, S.T. Deep learning-based annotation transfer between molecular imaging modalities: An automated workflow for multimodal data integration. Anal. Chem. 2021, 93, 3061–3071. [Google Scholar] [CrossRef]
- Pandiani, C.; Strub, T.; Nottet, N.; Cheli, Y.; Gambi, G.; Bille, K.; Husser, C.; Dalmasso, M.; Béranger, G.; Lassalle, S. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. 2021, 28, 1990–2000. [Google Scholar] [CrossRef]
- Petersen, D.; Naveed, P.; Ragheb, A.; Niedieker, D.; El-Mashtoly, S.; Brechmann, T.; Kötting, C.; Schmiegel, W.; Freier, E.; Pox, C. Raman fiber-optical method for colon cancer detection: Cross-validation and outlier identification approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 181, 270–275. [Google Scholar] [CrossRef]
- Hollon, T.C.; Pandian, B.; Adapa, A.R.; Urias, E.; Save, A.V.; Khalsa, S.S.S.; Eichberg, D.G.; D’Amico, R.S.; Farooq, Z.U.; Lewis, S. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 2020, 26, 52–58. [Google Scholar] [CrossRef]
- Phelps, D.L.; Balog, J.; Gildea, L.F.; Bodai, Z.; Savage, A.; El-Bahrawy, M.A.; Speller, A.V.; Rosini, F.; Kudo, H.; McKenzie, J.S. The surgical intelligent knife distinguishes normal, borderline and malignant gynaecological tissues using rapid evaporative ionisation mass spectrometry (REIMS). Br. J. Cancer 2018, 118, 1349–1358. [Google Scholar] [CrossRef]
- Römpp, A.; Schramm, T.; Hester, A.; Klinkert, I.; Both, J.-P.; Heeren, R.M.; Stoeckli, M.; Spengler, B. imzML: Imaging Mass Spectrometry Markup Language: A common data format for mass spectrometry imaging. In Data Mining in Proteomics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 205–224. [Google Scholar]
- Iakab, S.A.; Sementé, L.; García-Altares, M.; Correig, X.; Ràfols, P. Raman2imzML converts Raman imaging data into the standard mass spectrometry imaging format. BMC Bioinform. 2020, 21, 448. [Google Scholar] [CrossRef]
- Schapiro, D.; Yapp, C.; Sokolov, A.; Reynolds, S.M.; Chen, Y.-A.; Sudar, D.; Xie, Y.; Muhlich, J.; Arias-Camison, R.; Arena, S.; et al. MITI Minimum Information guidelines for highly multiplexed tissue images. Nat. Methods 2022, 19, 262–267. [Google Scholar] [CrossRef]
- Banas, A.; Banas, K.; Furgal-Borzych, A.; Kwiatek, W.M.; Pawlicki, B.; Breese, M. The pituitary gland under infrared light–in search of a representative spectrum for homogeneous regions. Analyst 2015, 140, 2156–2163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.; Momin, A.; Shaner, R.; Wang, E.; Bowen, N.J.; Matyunina, L.V.; Walker, L.; McDonald, J.F.; Sullards, M.C.; et al. Elevation of sulfatides in ovarian cancer: An integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol. Cancer 2010, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- ISO/IEC 17025:2005; General Requirements for the Competence of Testing and Calibration Laboratories. International Organization for Standardization: Vernier, Switzerland, 2005.
- Schneider, F.; Maurer, C.; Friedberg, R.C. International organization for standardization (ISO) 15189. Ann. Lab. Med. 2017, 37, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.; Emerson, P.; Pennington, G.; Lilleyman, J. Royal college of pathologists’ United kingdom pilot study of laboratory Accreditation. J. Clin. Pathol. 1990, 43, 89–91. [Google Scholar]
- Abu Amero, K. Overview of the laboratory accreditation programme of the College of American Pathologists. EMHJ-East. Mediterr. Health J. 2002, 8, 654–663. [Google Scholar] [CrossRef]
- Tzankov, A.; Tornillo, L. Hands-on experience: Accreditation of pathology laboratories according to ISO 15189. Pathobiology 2017, 84, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, R.J. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. J. Proteom. 2012, 75, 4893–4911. [Google Scholar] [CrossRef]
- Abdar, M.; Pourpanah, F.; Hussain, S.; Rezazadegan, D.; Liu, L.; Ghavamzadeh, M.; Fieguth, P.; Cao, X.; Khosravi, A.; Acharya, U.R. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf. Fusion 2021, 76, 243–297. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Gustafsson, O.J.; Winderbaum, L.J.; Condina, M.R.; Boughton, B.A.; Hamilton, B.R.; Undheim, E.A.; Becker, M.; Hoffmann, P. Balancing sufficiency and impact in reporting standards for mass spectrometry imaging experiments. Gigascience 2018, 7, giy102. [Google Scholar] [CrossRef]
- Chang, H.Y.; Jung, C.K.; Woo, J.I.; Lee, S.; Cho, J.; Kim, S.W.; Kwak, T.-Y. Artificial intelligence in pathology. J. Pathol. Transl. Med. 2019, 53, 1. [Google Scholar] [PubMed] [Green Version]

| Technique | Measurements | Specificity | Sensitivity | Spatial Resolution | Labelled | Spectral Channels |
|---|---|---|---|---|---|---|
| Raman microscopy | Bond vibration | Medium | Medium | <1 μm | Unlabelled | 100–1000 |
| IR microscopy | Bond vibration | Medium | Medium | 1 μm | Unlabelled | 100–1000 |
| Hyperspectral imaging | Light absorption | Low | Low | 100 nm | Unlabelled | 10–100 |
| Multiplex IHC | Labelled antibodies | High | High | 100 nm | Labelled | 10 |
| Multiplex FISH | Labelled fluorophores | High | High | 100 nm | Labelled | 10 |
| IMC | Labelled antibodies | High | High | 1 μm | Labelled | 52 |
| MSI | Ionised molecules | High | Medium | 10 μm | Unlabelled | 1000–100,000 |
| Microdissection and RNA-seq | Gene expression | High | High | 1 μm | Unlabelled | 1000–100,000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dexter, A.; Tsikritsis, D.; Belsey, N.A.; Thomas, S.A.; Venton, J.; Bunch, J.; Romanchikova, M. Next Generation Digital Pathology: Emerging Trends and Measurement Challenges for Molecular Pathology. J. Mol. Pathol. 2022, 3, 168-181. https://doi.org/10.3390/jmp3030014
Dexter A, Tsikritsis D, Belsey NA, Thomas SA, Venton J, Bunch J, Romanchikova M. Next Generation Digital Pathology: Emerging Trends and Measurement Challenges for Molecular Pathology. Journal of Molecular Pathology. 2022; 3(3):168-181. https://doi.org/10.3390/jmp3030014
Chicago/Turabian StyleDexter, Alex, Dimitrios Tsikritsis, Natalie A. Belsey, Spencer A. Thomas, Jenny Venton, Josephine Bunch, and Marina Romanchikova. 2022. "Next Generation Digital Pathology: Emerging Trends and Measurement Challenges for Molecular Pathology" Journal of Molecular Pathology 3, no. 3: 168-181. https://doi.org/10.3390/jmp3030014
APA StyleDexter, A., Tsikritsis, D., Belsey, N. A., Thomas, S. A., Venton, J., Bunch, J., & Romanchikova, M. (2022). Next Generation Digital Pathology: Emerging Trends and Measurement Challenges for Molecular Pathology. Journal of Molecular Pathology, 3(3), 168-181. https://doi.org/10.3390/jmp3030014






