Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective
Abstract
1. Introduction
2. Which Biomarker Tests Are Currently Recommended in Europe for NSCLC-Targeted Treatment?
3. How Many Patients Miss out on Biomarker Testing/Molecular Diagnosis in Europe?
4. Why Are Patients Missing out on Biomarker Testing/Molecular Diagnosis?
5. What Is Liquid Biopsy?
6. What Are the Current Recommendations for Liquid Biopsy in Europe?
7. Beyond the Guidelines: What Does Liquid Biopsy and NGS Use in Europe Actually Look Like?
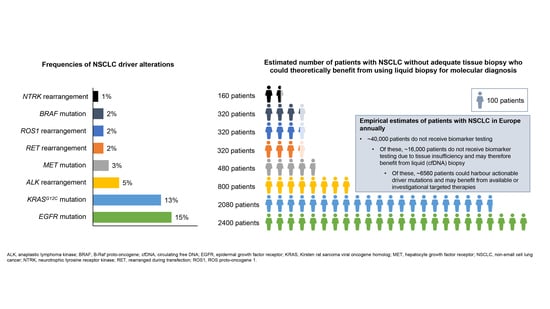
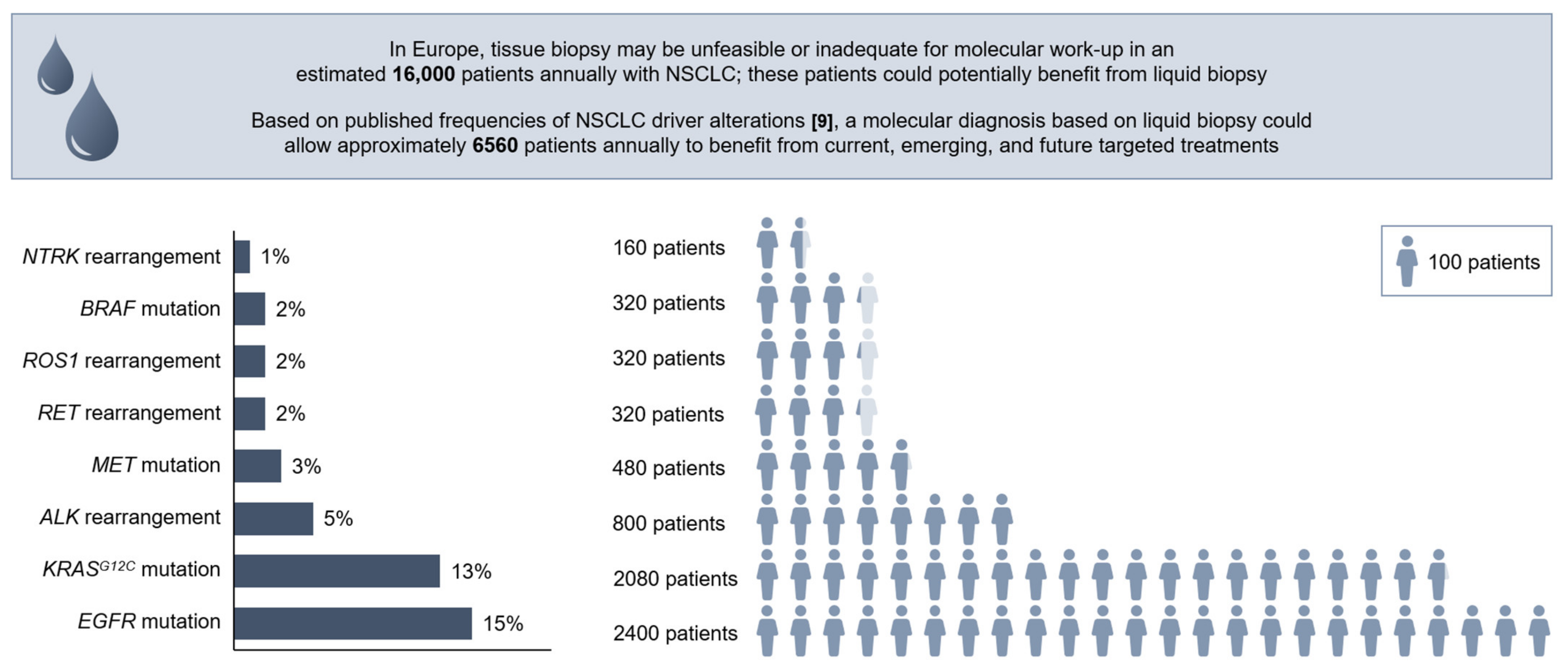
8. The Potential for Broader Adoption of Liquid Biopsy
9. What Might Be Next for Liquid Biopsy in NSCLC? An Expert Opinion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Respiratory Society. ERS White Book. Chapter 19: Lung Cancer. Available online: https://www.erswhitebook.org/files/public/Chapters/19_lung_cancer.pdf (accessed on 8 June 2021).
- European Society for Medical Oncology. Metastatic Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Available online: https://www.esmo.org/content/download/347819/6934778/1/ESMO-CPG-mNSCLC-15SEPT2020.pdf (accessed on 8 June 2021).
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. The IASLC Lung Cancer Staging Project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef]
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The effect of advances in lung-cancer treatment on population mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Griesinger, F.; Eberhardt, W.; Nusch, A.; Reiser, M.; Zahn, M.O.; Maintz, C.; Bernhardt, C.; Losem, C.; Stenzinger, A.; Heukamp, L.C.; et al. Biomarker testing in non-small cell lung cancer in routine care: Analysis of the first 3717 patients in the German prospective, observational, nation-wide CRISP Registry (AIO-TRK-0315). Lung Cancer 2021, 152, 174–184. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Tarceva Summary of Product Characteristics. Available online: https://www.ema.europa.eu/en/documents/product-information/tarceva-epar-product-information_en.pdf (accessed on 8 June 2021).
- Kerr, K.M.; Bibeau, F.; Thunnissen, E.; Botling, J.; Ryška, A.; Wolf, J.; Öhrling, K.; Burdon, P.; Malapelle, U.; Büttner, R. The evolving landscape of biomarker testing for non-small cell lung cancer in Europe. Lung Cancer 2021, 154, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. FDA Approves First Targeted Therapy for Lung Cancer Mutation Previously Considered Resistant to Drug Therapy. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-lung-cancer-mutation-previously-considered-resistant-drug (accessed on 8 June 2021).
- US Food and Drug Administration. FDA Approves First Targeted Therapy for Subset of Non-Small Cell Lung Cancer. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-subset-non-small-cell-lung-cancer (accessed on 22 June 2021).
- Smeltzer, M.P.; Wynes, M.W.; Lantuejoul, S.; Soo, R.; Ramalingam, S.S.; Varella-Garcia, M.; Meadows Taylor, M.; Richeimer, K.; Wood, K.; Howell, K.E.; et al. The International Association for the Study of Lung Cancer global survey on molecular testing in lung cancer. J. Thorac. Oncol. 2020, 15, 1434–1448. [Google Scholar] [CrossRef]
- Volckmar, A.L.; Leichsenring, J.; Kirchner, M.; Christopoulos, P.; Neumann, O.; Budczies, J.; Morais de Oliveira, C.M.; Rempel, E.; Buchhalter, I.; Brandt, R.; et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3,000 Heidelberg cases. Int. J. Cancer 2019, 145, 649–661. [Google Scholar] [CrossRef]
- Pruneri, G.; De Braud, F.; Sapino, A.; Aglietta, M.; Vecchione, A.; Giusti, R.; Marchio, C.; Scarpino, S.; Baggi, A.; Bonetti, G.; et al. Next-generation sequencing in clinical practice: Is it a cost-saving alternative to a single-gene testing approach? Pharmacoecon. Open 2021, 5, 285–298. [Google Scholar] [CrossRef]
- Dall’Olio, F.G.; Conci, N.; Rossi, G.; Fiorentino, M.; De Giglio, A.; Grilli, G.; Altimari, A.; Gruppioni, E.; Filippini, D.M.; Di Federico, A.; et al. Comparison of sequential testing and next generation sequencing in advanced lung adenocarcinoma patients-A single centre experience. Lung Cancer 2020, 149, 5–9. [Google Scholar] [CrossRef]
- Baggi, A.; Bonetti, G.; Gancitano, G.; Scalamogna, R.; Peccerillo, C.; Volpe, M.; Franzini, J.M.; Vecchione, A.; Sapino, A.; Pruneri, G.; et al. Organizational and economic impact of next generation sequencing and hotspot approach. Value Health 2019, 22, S470. [Google Scholar] [CrossRef]
- Remon, J.; Swalduz, A.; Planchard, D.; Ortiz-Cuaran, S.; Mezquita, L.; Lacroix, L.; Jovelet, C.; Rouleau, E.; Leonce, C.; De Kievit, F.; et al. Outcomes in oncogenic-addicted advanced NSCLC patients with actionable mutations identified by liquid biopsy genomic profiling using a tagged amplicon-based NGS assay. PLoS ONE 2020, 15, e0234302. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef]
- Kim, S.T.; Banks, K.C.; Lee, S.H.; Kim, K.; Park, J.O.; Park, S.H.; Park, Y.S.; Lim, H.Y.; Kang, W.K.; Lanman, R.B.; et al. Prospective feasibility study for using cell-free circulating tumor DNA-guided therapy in refractory metastatic solid cancers: An interim analysis. JCO Precis. Oncol. 2017, 1, 1–15. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Non-Small Cell Lung Cancer. Version 5.2021. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450 (accessed on 13 August 2021).
- Lee, D.H.; Tsao, M.S.; Kambartel, K.O.; Isobe, H.; Huang, M.S.; Barrios, C.H.; Khattak, A.; de Marinis, F.; Kothari, S.; Arunachalam, A.; et al. Molecular testing and treatment patterns for patients with advanced non-small cell lung cancer: PIvOTAL observational study. PLoS ONE 2018, 13, e0202865. [Google Scholar] [CrossRef] [PubMed]
- Salas, C.; Martin-Lopez, J.; Martinez-Pozo, A.; Hernandez-Iglesias, T.; Carcedo, D.; Ruiz de Alda, L.; Garcia, J.F.; Rojo, F. Real-world biomarker testing rate and positivity rate in NSCLC in Spain: Prospective Central Lung Cancer Biomarker Testing Registry (LungPath) from the Spanish Society of Pathology (SEAP). J. Clin. Pathol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Robert, N.J.; Nwokeji, E.D.; Espirito, J.L.; Chen, L.; Karhade, M.; Evangelist, M.C.; Spira, A.I.; Neubauer, M.A.; Bullock, S.A.; Coleman, R.L.; et al. Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J. Clin. Oncol. 2021, 39, 9004. [Google Scholar] [CrossRef]
- Chen, V.W.; Ruiz, B.A.; Hsieh, M.C.; Wu, X.C.; Ries, L.A.; Lewis, D.R. Analysis of stage and clinical/prognostic factors for lung cancer from SEER registries: AJCC staging and collaborative stage data collection system. Cancer 2014, 120, 3781–3792. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: The lung cancer mutation consortium experience. J. Thorac. Oncol. 2015, 10, 768–777. [Google Scholar] [CrossRef]
- Bosc, C.; Ferretti, G.R.; Cadranel, J.; Audigier-Valette, C.; Besse, B.; Barlesi, F.; Decroisette, C.; Lantuejoul, S.; Arbib, F.; Moro-Sibilot, D. Rebiopsy during disease progression in patients treated by TKI for oncogene-addicted NSCLC. Target. Oncol. 2015, 10, 247–253. [Google Scholar] [CrossRef]
- Boskovic, T.; Stanic, J.; Pena-Karan, S.; Zarogoulidis, P.; Drevelegas, K.; Katsikogiannis, N.; Machairiotis, N.; Mpakas, A.; Tsakiridis, K.; Kesisis, G.; et al. Pneumothorax after transthoracic needle biopsy of lung lesions under CT guidance. J. Thorac. Dis. 2014, 6, S99–S107. [Google Scholar] [CrossRef]
- Tredan, O.; Wang, Q.; Pissaloux, D.; Cassier, P.; de la Fouchardiere, A.; Fayette, J.; Desseigne, F.; Ray-Coquard, I.; de la Fouchardiere, C.; Frappaz, D.; et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: Analysis from the ProfiLER trial. Ann. Oncol. 2019, 30, 757–765. [Google Scholar] [CrossRef]
- Murray, S.; Karavasilis, V.; Bobos, M.; Razis, E.; Papadopoulos, S.; Christodoulou, C.; Kosmidis, P.; Fountzilas, G. Molecular predictors of response to tyrosine kinase inhibitors in patients with non-small-cell lung cancer. J. Exp. Clin. Cancer Res. 2012, 31, 77. [Google Scholar] [CrossRef] [PubMed]
- Chouaid, C.; Dujon, C.; Do, P.; Monnet, I.; Madroszyk, A.; Le Caer, H.; Auliac, J.B.; Berard, H.; Thomas, P.; Lena, H.; et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: A prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014, 86, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Tsao, M.S.; Le, L.W.; Shepherd, F.A.; Feld, R.; Burkes, R.L.; Liu, G.; Kamel-Reid, S.; Hwang, D.; Tanguay, J.; et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 2015, 26, 1415–1421. [Google Scholar] [CrossRef]
- Gutierrez, M.E.; Choi, K.; Lanman, R.B.; Licitra, E.J.; Skrzypczak, S.M.; Pe Benito, R.; Wu, T.; Arunajadai, S.; Kaur, S.; Harper, H.; et al. Genomic profiling of advanced non-small cell lung cancer in community settings: Gaps and opportunities. Clin. Lung Cancer 2017, 18, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Agha, A.; Devost, N.; Walton, R.N.; Ghosh, S.; Ho, C. Biopsy on progression in patients with EGFR mutation-positive advanced non-small-cell lung cancer-a Canadian experience. Curr. Oncol. 2020, 27, 27–33. [Google Scholar] [CrossRef]
- Ofiara, L.M.; Navasakulpong, A.; Beaudoin, S.; Gonzalez, A.V. Optimizing tissue sampling for the diagnosis, subtyping, and molecular analysis of lung cancer. Front. Oncol. 2014, 4, 253. [Google Scholar] [CrossRef]
- Yoon, H.J.; Lee, H.Y.; Lee, K.S.; Choi, Y.L.; Ahn, M.J.; Park, K.; Ahn, J.S.; Sun, J.M.; Kim, J.; Kim, T.S.; et al. Repeat biopsy for mutational analysis of non-small cell lung cancers resistant to previous chemotherapy: Adequacy and complications. Radiology 2012, 265, 939–948. [Google Scholar] [CrossRef]
- Hong, M.H.; Kim, H.R.; Ahn, B.C.; Heo, S.J.; Kim, J.H.; Cho, B.C. Real-world analysis of the efficacy of rebiopsy and EGFR mutation test of tissue and plasma samples in drug-resistant non-small cell lung cancer. Yonsei Med. J. 2019, 60, 525–534. [Google Scholar] [CrossRef]
- Tateishi, A.; Matsumoto, Y.; Tanaka, M.; Nakai, T.; Sasada, S.; Aoshima, M.; Tsuchida, T. The utility of transbronchial rebiopsy for peripheral pulmonary lesions in patients with advanced non-squamous non-small cell lung cancer. BMC Pulm. Med. 2020, 20, 238. [Google Scholar] [CrossRef]
- Tsai, E.B.; Pomykala, K.; Ruchalski, K.; Genshaft, S.; Abtin, F.; Gutierrez, A.; Kim, H.J.; Li, A.; Adame, C.; Jalalian, A.; et al. Feasibility and safety of intrathoracic biopsy and repeat biopsy for evaluation of programmed cell death ligand-1 expression for immunotherapy in non-small cell lung cancer. Radiology 2018, 287, 326–332. [Google Scholar] [CrossRef]
- Haentschel, M.; Boeckeler, M.; Bonzheim, I.; Schimmele, F.; Spengler, W.; Stanzel, F.; Petermann, C.; Darwiche, K.; Hagmeyer, L.; Buettner, R.; et al. Influence of biopsy technique on molecular genetic tumor characterization in non-small cell lung cancer-the prospective, randomized, single-blinded, multicenter PROFILER study protocol. Diagnostics 2020, 10, 459. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid biopsy for advanced non-small cell lung cancer: A consensus statement from the International Association for the Study of Lung Cancer (IASLC). J. Thorac. Oncol. 2021. [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): A statement paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Muller, J.N.; Falk, M.; Talwar, J.; Neemann, N.; Mariotti, E.; Bertrand, M.; Zacherle, T.; Lakis, S.; Menon, R.; Gloeckner, C.; et al. Concordance between comprehensive cancer genome profiling in plasma and tumor specimens. J. Thorac. Oncol. 2017, 12, 1503–1511. [Google Scholar] [CrossRef]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Janne, P.A.; Oxnard, G.R. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016, 2, 1014–1022. [Google Scholar] [CrossRef]
- Rolfo, C.; Cardona, A.F.; Cristofanilli, M.; Paz-Ares, L.; Diaz Mochon, J.J.; Duran, I.; Raez, L.E.; Russo, A.; Lorente, J.A.; Malapelle, U.; et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit. Rev. Oncol. Hematol. 2020, 151, 102978. [Google Scholar] [CrossRef]
- Cho, M.S.; Park, C.H.; Lee, S.; Park, H.S. Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS ONE 2020, 15, e0230622. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Lianidou, E.; Pantel, K. Liquid biopsies. Genes Chromosomes Cancer 2019, 58, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Esposito Abate, R.; Frezzetti, D.; Maiello, M.R.; Gallo, M.; Camerlingo, R.; De Luca, A.; De Cecio, R.; Morabito, A.; Normanno, N. Next generation sequencing-based profiling of cell free DNA in patients with advanced non-small cell lung cancer: Advantages and pitfalls. Cancers 2020, 12, 3804. [Google Scholar] [CrossRef]
- Mograbi, B.; Heeke, S.; Hofman, P. The Importance of STK11/LKB1 assessment in non-small cell lung carcinomas. Diagnostics 2021, 11, 196. [Google Scholar] [CrossRef]
- Del Re, M.; Rofi, E.; Cappelli, C.; Puppo, G.; Crucitta, S.; Valeggi, S.; Chella, A.; Danesi, R.; Petrini, I. The increase in activating EGFR mutation in plasma is an early biomarker to monitor response to osimertinib: A case report. BMC Cancer 2019, 19, 410. [Google Scholar] [CrossRef] [PubMed]
- National Institute for Health and Care Excellence. Plasma EGFR Mutation Tests for Adults with Locally Advanced or Metastatic Non-Small-Cell Lung Cancer [Medtech Innovation Briefing MIB137]. Available online: https://www.nice.org.uk/advice/mib137 (accessed on 8 June 2021).
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Chan, H.T.; Chin, Y.M.; Nakamura, Y.; Low, S.K. Clonal hematopoiesis in liquid biopsy: From biological noise to valuable clinical implications. Cancers 2020, 12, 2277. [Google Scholar] [CrossRef]
- Dorantes-Heredia, R.; Ruiz-Morales, J.M.; Cano-Garcia, F. Histopathological transformation to small-cell lung carcinoma in non-small cell lung carcinoma tumors. Transl. Lung Cancer Res. 2016, 5, 401–412. [Google Scholar] [CrossRef]
- Herbreteau, G.; Vallee, A.; Charpentier, S.; Normanno, N.; Hofman, P.; Denis, M.G. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): Clinical application and future perspectives. J. Thorac. Dis. 2019, 11, S113–S126. [Google Scholar] [CrossRef]
- Thalanayar, P.M.; Altintas, N.; Weissfeld, J.L.; Fuhrman, C.R.; Wilson, D.O. Indolent, potentially inconsequential lung cancers in the Pittsburgh Lung Screening Study. Ann. Am. Thorac. Soc. 2015, 12, 1193–1196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- Bauml, J.; Levy, B. Clonal hematopoiesis: A new layer in the liquid biopsy story in lung cancer. Clin. Cancer Res. 2018, 24, 4352–4354. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ulrich, B.C.; Supplee, J.; Kuang, Y.; Lizotte, P.H.; Feeney, N.B.; Guibert, N.M.; Awad, M.M.; Wong, K.K.; Janne, P.A.; et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin. Cancer Res. 2018, 24, 4437–4443. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Costa, J.L.; Pepe, F.; Russo, G.; Gragnano, G.; Russo, A.; Iaccarino, A.; de Miguel-Perez, D.; Serrano, M.J.; Denninghoff, V.; et al. Next generation sequencing for liquid biopsy based testing in non-small cell lung cancer in 2021. Crit. Rev. Oncol. Hematol. 2021, 161, 103311. [Google Scholar] [CrossRef]
- Foundation Medicine. FDA Approves Foundation Medicine’s FoundationOne® Liquid CDx, a Comprehensive Pan-Tumor Liquid Biopsy Test with Multiple Companion Diagnostic Indications for Patients with Advanced Cancer. Available online: https://www.foundationmedicine.com/press-releases/445c1f9e-6cbb-488b-84ad-5f133612b721 (accessed on 8 June 2021).
- US Food and Drug Administration. FDA Approves Pralsetinib for Lung Cancer with RET Gene Fusions. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-pralsetinib-lung-cancer-ret-gene-fusions (accessed on 8 June 2021).
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 129–159. [Google Scholar] [CrossRef]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Chugai Pharmaceutical. Chugai Obtains Approval for FoundationOne Liquid CDx Cancer Genomic Profile, the First Blood-Based Comprehensive Genomic Profiling Test for Solid Tumors in Japan. Available online: https://www.chugai-pharm.co.jp/english/news/detail/20210323170004_807.html (accessed on 8 June 2021).
- CISION PR Newswire. Guardant360 CDx Submitted for Regulatory Approval in Japan. Available online: https://www.prnewswire.com/news-releases/guardant360-cdx-submitted-for-regulatory-approval-in-japan-301221090.html (accessed on 8 June 2021).
- Gobierno de Espana: Ministerio de Ciencia, Innovacion y Universidades; Junta de Andalucia; FEDER. Final Report on the Meeting of Experts External for the Preparation of a Document Consensus on Development Priorities in Biopsy Liquid. Available online: https://www.sspa.juntadeandalucia.es/servicioandaluzdesalud/sites/default/files/sincfiles/wsas-media-mediafile_sasdocumento/2019/CONSENSO%20EXP%20EXT_%20LB_%20GENYO%2014%20JUN%202018_revisado_completo_edit_f.pdf (accessed on 8 June 2021).
- Beretta, G.; Capoluongo, E.; Danesi, R.; Del Re, M.; Fassan, M.; Giuffrè, G.; Gori, S.; Gristina, V.; Incorvaia, L.; Malapelle, U.; et al. 2020 Recommendations for Performing Molecular Tests on Liquid Biopsy in Oncology. Available online: https://www.aiom.it/wp-content/uploads/2020/07/2020_Raccomandazioni_Biopsia_Liquida.pdf (accessed on 8 June 2021).
- Guibert, N.; Pradines, A.; Favre, G.; Mazieres, J. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur. Respir. Rev. 2020, 29, 190052. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Garcia-Campelo, R.; de Alava, E.; Vera, R.; Rodriguez-Peralto, J.L.; Rodriguez-Lescure, A.; Bellosillo, B.; Garrido, P.; Rojo, F.; Alvarez-Alegret, R. Liquid biopsy in oncology: A consensus statement of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin. Transl. Oncol. 2020, 22, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Sirera, R.; Jantus-Lewintre, E.; Reclusa, P.; Calabuig-Farinas, S.; Blasco, A.; Pisapia, P.; Rolfo, C.; Camps, C. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev. Mol. Diagn. 2017, 17, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, 9650. [Google Scholar] [CrossRef]
- Jovelet, C.; Madic, J.; Remon, J.; Honore, A.; Girard, R.; Rouleau, E.; Andre, B.; Besse, B.; Droniou, M.; Lacroix, L. Crystal digital droplet PCR for detection and quantification of circulating EGFR sensitizing and resistance mutations in advanced non-small cell lung cancer. PLoS ONE 2017, 12, e0183319. [Google Scholar] [CrossRef]
- Vessies, D.C.L.; Greuter, M.J.E.; van Rooijen, K.L.; Linders, T.C.; Lanfermeijer, M.; Ramkisoensing, K.L.; Meijer, G.A.; Koopman, M.; Coupe, V.M.H.; Vink, G.R.; et al. Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing. Sci. Rep. 2020, 10, 8122. [Google Scholar] [CrossRef]
- Pakkala, S.; Ramalingam, S.S. Personalized therapy for lung cancer: Striking a moving target. JCI Insight 2018, 3, e120858. [Google Scholar] [CrossRef]
- Skov, B.G.; Rorvig, S.B.; Jensen, T.H.L.; Skov, T. The prevalence of programmed death ligand-1 (PD-L1) expression in non-small cell lung cancer in an unselected, consecutive population. Mod. Pathol. 2020, 33, 109–117. [Google Scholar] [CrossRef]
- Nassar, A.H.; Adib, E.; Kwiatkowski, D.J. Distribution of KRAS (G12C) somatic mutations across race, sex, and cancer type. N. Engl. J. Med. 2021, 384, 185–187. [Google Scholar] [CrossRef]
- Huang, K.L.; Wang, S.Y.; Lu, W.C.; Chang, Y.H.; Su, J.; Lu, Y.T. Effects of low-dose computed tomography on lung cancer screening: A systematic review, meta-analysis, and trial sequential analysis. BMC Pulm. Med. 2019, 19, 126. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Maddala, T.; Hubbell, E.; Aravanis, A.; Zhang, N.; Venn, O.; Valouev, A.; Shen, L.; Patel, S.; Jamshidi, A.; et al. Genome-wide sequencing for early stage lung cancer detection from plasma cell-free DNA (cfDNA): The Circulating Cancer Genome Atlas (CCGA) study. J. Clin. Oncol. 2018, 36, LBA8501. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Cree, I.A.; Uttley, L.; Buckley Woods, H.; Kikuchi, H.; Reiman, A.; Harnan, S.; Whiteman, B.L.; Philips, S.T.; Messenger, M.; Cox, A.; et al. The evidence base for circulating tumour DNA blood-based biomarkers for the early detection of cancer: A systematic mapping review. BMC Cancer 2017, 17, 697. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Siravegna, G. How to use liquid biopsies to treat patients with cancer. ESMO Open 2021, 6, 100060. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.J.; Garrido-Navas, M.C.; Diaz Mochon, J.J.; Cristofanilli, M.; Gil-Bazo, I.; Pauwels, P.; Malapelle, U.; Russo, A.; Lorente, J.A.; Ruiz-Rodriguez, A.J.; et al. Precision prevention and cancer interception: The new challenges of liquid biopsy. Cancer Discov. 2020, 10, 1635–1644. [Google Scholar] [CrossRef]
- Bordi, P.; Del Re, M.; Minari, R.; Rofi, E.; Buti, S.; Restante, G.; Squadrilli, A.; Crucitta, S.; Casartelli, C.; Gnetti, L.; et al. From the beginning to resistance: Study of plasma monitoring and resistance mechanisms in a cohort of patients treated with osimertinib for advanced T790M-positive NSCLC. Lung Cancer 2019, 131, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Ishiba, T.; Hoffmann, A.C.; Usher, J.; Elshimali, Y.; Sturdevant, T.; Dang, M.; Jaimes, Y.; Tyagi, R.; Gonzales, R.; Grino, M.; et al. Frequencies and expression levels of programmed death ligand 1 (PD-L1) in circulating tumor RNA (ctRNA) in various cancer types. Biochem. Biophys. Res. Commun. 2018, 500, 621–625. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Lam, S.N.; Zhou, Y.C.; Chan, Y.M.; Foo, C.M.; Lee, P.Y.; Mok, W.Y.; Wong, W.S.; Fung, Y.Y.; Wong, K.Y.; Huang, J.Y.; et al. Comparison of target enrichment platforms for circulating tumor DNA detection. Sci. Rep. 2020, 10, 4124. [Google Scholar] [CrossRef] [PubMed]
- Vendrell, J.A.; Quantin, X.; Serre, I.; Solassol, J. Combination of tissue and liquid biopsy molecular profiling to detect transformation to small cell lung carcinoma during osimertinib treatment. Ther. Adv. Med. Oncol. 2020, 12, 1758835920974192. [Google Scholar] [CrossRef] [PubMed]
- Lebofsky, R.; Decraene, C.; Bernard, V.; Kamal, M.; Blin, A.; Leroy, Q.; Rio Frio, T.; Pierron, G.; Callens, C.; Bieche, I.; et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol. Oncol. 2015, 9, 783–790. [Google Scholar] [CrossRef] [PubMed]


| Gene | Alteration | Prevalence | ESCAT |
|---|---|---|---|
| Level I alterations: “The ESMO Precision Medicine Working Group recommends that a tumour (or plasma) sample from a patient with advanced non-squamous NSCLC is profiled using NGS technology in order to detect level I alterations” | |||
| EGFR | Common mutations (Del19, L858R) Acquired T790M exon 20 Uncommon EGFR mutations (G719X in exon 18, L861Q in exon 21, S768I in exon 20) | 15% (50–60% in Asians) 60% of EGFR mutant NSCLC10% | IA IA IB |
| ALK | Fusions (mutations as mechanism of resistance) | 5% | IA |
| MET | Mutations ex 14 skipping | 3% | IB |
| BRAFV600E | Mutations | 2% | IB |
| ROS1 | Fusions (mutations as mechanism of resistance) | 1–2% | IB |
| NTRK | Fusions | 0.23–3% | IC |
| RET | Fusions | 1–2% | IC |
| Level II–III alterations: “There is no evidence that panels detecting genes with a lower level of evidence brings additional value from a public health perspective. They could be used only if the report ranks genomic alterations according to valid ranking systems (e.g., ESCAT, OncoKB) and on the basis of specific agreements with payers taking into account the overall cost of the strategy (including off label use of drugs) as compared with small panels.” | |||
| EGFR | Exon 20 insertions | 2% | IIB |
| MET | Focal amplifications (acquired resistance on EGFR TKI in EGFR-mutant tumours) | 3% | IIB |
| KRASG12C | Mutations | 12% | IIB |
| ERBB2/HER2 | Hotspot mutations Amplifications | 2–5% | IIB |
| BRCA 1/2 | Mutations | 1.2% | IIIA |
| PIK3CA | Hotspot mutations | 1.2–7% | IIIA |
| NRG1 | Fusions | 1.7% | IIIB |
| Article | Summary | Biopsy Not Possible/ Not Carried Out (%) | Biopsy Sample Inadequate (%) | Overall Biopsy Failure Rate (%) |
|---|---|---|---|---|
| [22] (Italy, Spain, and Germany) | A chart review study including 515 patients in Italy, Spain, and Germany with advanced newly diagnosed NSCLC; 468/505 had a biopsy. | IB: 9% | NR | NR |
| [29] (France) | A prospective study of 2579 patients with advanced cancer (6% lung cancer) who progressed after 1 L treatment and were potentially eligible for molecular-based therapies. A total of 435 patients (17%) were withdrawn from the study (insufficient quantity or quality of tumour sample, n = 357; tumour sample < 10% tumour cells, n = 19; DNA extraction/quantity issues, n = 19). | NR | 17% | 17% |
| [30] (Greece) | A retrospective study of 72 patients with histologically confirmed advanced/metastatic NSCLC who received anti-EGFR TKIs (any line); most (56%) received 2L treatment. Five patients were excluded (7.8%) due to insufficient tumour in biopsies. | NR | 7.8% | 7.8% |
| [27] (France) | A retrospective study of 84 patients with lung cancer (93% adenocarcinoma) with documented EGFR mutation or ALK rearrangement who developed radiographic progression on TKIs. Thirty-nine patients (46%) underwent re-biopsy at the time of acquired resistance. Among the 39 re-biopsies, there was sufficient tissue for histopathological or cytological examination in 89.7% of cases; in three cases there was no tumour tissue, and one case showed necrotic tissue. Re-biopsy was considered feasible in 33 of 45 patients (73%) who did not undergo re-biopsy. | RB: 14% | RB: 10% | 19% |
| [31] (France) | A prospective study of 100 patients with advanced NSCLC with RECIST-defined progression after 1 L therapy and a clinical indication for re-biopsy. Re-biopsy was not feasible in 18% of cases. Of the 82 patients who underwent re-biopsy, 94% could be analysed histologically. Upon histological examination, 18.3% of samples contained no tumour cells, and 7.3% of samples contained too few tumour cells for molecular analysis. | RB: 18% | RB: 26% | 43% |
| Limitations and Considerations | Details |
| Test sensitivity |
|
| Potential for false positives | |
| Morphological transformation | |
| Challenges in detecting certain types of gene fusion translocations or splice variant alterations |
|
| Detection of germline variants |
|
| Guideline | Recommendations on Liquid Biopsy for Molecular Diagnosis | Recommendations on Multiplex Panel versus Single-Gene Testing (Recommendations Specific to Liquid Biopsy Are Bolded) |
|---|---|---|
| ESMO NSCLC [2] |
|
|
| ESMO PMWG [10] |
|
|
| NCCN [21] |
|
|
| CAP/IASLC/AMP and ASCO [69] |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malapelle, U.; Tiseo, M.; Vivancos, A.; Kapp, J.; Serrano, M.J.; Tiemann, M. Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective. J. Mol. Pathol. 2021, 2, 255-273. https://doi.org/10.3390/jmp2030022
Malapelle U, Tiseo M, Vivancos A, Kapp J, Serrano MJ, Tiemann M. Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective. Journal of Molecular Pathology. 2021; 2(3):255-273. https://doi.org/10.3390/jmp2030022
Chicago/Turabian StyleMalapelle, Umberto, Marcello Tiseo, Ana Vivancos, Joshua Kapp, M. Josè Serrano, and Markus Tiemann. 2021. "Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective" Journal of Molecular Pathology 2, no. 3: 255-273. https://doi.org/10.3390/jmp2030022
APA StyleMalapelle, U., Tiseo, M., Vivancos, A., Kapp, J., Serrano, M. J., & Tiemann, M. (2021). Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective. Journal of Molecular Pathology, 2(3), 255-273. https://doi.org/10.3390/jmp2030022