Abstract
The past decade has witnessed significant advances in the application of molecular diagnostics for the pre-operative risk-stratification of cytologically indeterminate thyroid nodules. The tests that are currently marketed in the United States for this purpose combine aspects of tumor genotyping with gene and/or microRNA expression profiling. This review compares the general methodology and clinical validation studies for the three tests currently offered in the United States: ThyroSeq v3, Afirma GSC and Xpression Atlas, and ThyGeNEXT/ThyraMIR.
1. Background
Thyroid nodules are a common clinical finding, with palpable thyroid nodules detected in 1% of men and 5% of women in iodine-sufficient regions of the world [1,2,3,4,5]. However, with the increasing use of radiologic screening, one or more thyroid nodules are found in 60–70% of the population [1,6]. Most thyroid nodules (>90%) are asymptomatic and benign; of those that are malignant, the majority are low-risk neoplasms that do not alter survival. The current management and malignancy risk assessment of thyroid nodules involves a synthesis of clinical findings, ultrasound evaluation, and fine-needle aspiration (FNA) [1,7]. Specific ultrasound characteristics of thyroid tumors, such as hypoechogenicity, taller-than-wide shape on transverse view, irregular margins, microcalcifications/echogenic foci, and apparent extrathyroidal extension are associated with malignancy. These and other ultrasound features have been employed by various professional societies to develop ultrasound risk assessment schemes to select thyroid nodules for FNA biopsy [1,8,9,10].
FNA has proven to be an important tool in the management of thyroid nodules. Currently, thyroid FNA cytology is reported in a standardized fashion by employing one of the known tiered classification systems [3,11,12,13,14]. While FNA cytology can effectively differentiate between benign and malignant thyroid lesions in up to 75% of cases, 20–30% of aspirated nodules are reported as indeterminate for malignancy [14,15]. Each of the thyroid FNA classification schemes includes diagnostic categories to facilitate reporting of thyroid nodules that lack clearly defined cytomorphologic features of malignancy (Table 1) [14]. Some authors have suggested inclusion of the “Suspicious for Malignancy” interpretive category among indeterminate cases; however, the relatively high cancer risk and different management recommendations for nodules classified as “Suspicious for Malignancy” have prompted other authors to consider this category separately from the lower-risk indeterminate categories [4,14,16]. In general, the risk of malignancy for nodules in the lower-risk indeterminate categories ranges from 10 to 40%. This wide range of malignancy risk likely reflects variations in (1) the prevalence of thyroid cancer among different patient populations, (2) radiologic criteria for selecting nodules for FNA biopsy, (3) cytopathologists’ thresholds for classifying aspirates as indeterminate, and (4) surgical pathologists’ diagnostic thresholds for classifying thyroid nodules on thyroidectomy specimens. In spite of the challenges raised by indeterminate thyroid cytology diagnoses, both clinicians and pathologists generally agree that reporting of thyroid FNA cytology using standardized, tiered classification schemes is an important component of patient management [2,17].

Table 1.
Classification schemes for reporting thyroid cytology. Indeterminate diagnostic categories are shaded.
In recent years, advances in the clinical, pathologic, and molecular characterization of thyroid neoplasms have had major impacts on thyroid nodule management. An example of such a development is the establishment of strict criteria for reclassifying a subset of tumors that were formerly considered malignant (i.e., non-invasive, encapsulated follicular variant of papillary carcinoma) as indolent neoplasms (non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP)) [18,19,20,21]. This nomenclature change from “carcinoma” to “neoplasm” has reduced the cancer risk estimates associated with both the lower-risk indeterminate and “Suspicious for Malignancy” categories of thyroid FNA reporting systems [22,23]. Additionally, progress in understanding the molecular pathogenesis of thyroid neoplasms has fueled the development of molecular diagnostic tests for improving the preoperative risk-stratification of cytologically indeterminate thyroid nodules [24]. The primary goal of these molecular tests is to correctly identify benign nodules among those with indeterminate cytology, thereby decreasing the number of unnecessary diagnostic surgeries [25,26,27,28]. The updated versions of thyroid FNA classification schemes are now aligned with the aforementioned changes in histopathologic classification of thyroid neoplasms, applicability of molecular testing, and current recommendations for management.
2. Molecular Profiles of Thyroid Tumors
For the purposes of ancillary testing, the molecular changes that have been described in thyroid tumors may be conceptualized as those that occur on the DNA level versus those that manifest as downstream alterations in gene and/or microRNA expression profiles.
2.1. DNA Sequence Alterations, Gene Fusions, and Chromosomal Copy-Number Alterations
Genomic characterizations of various types of thyroid cancer have identified recurrent somatic mutations (single nucleotide variants, insertions, deletions) and gene fusions in hotspots within oncogenes and tumor suppressor genes as well as large-scale chromosomal copy number alterations [29,30,31,32,33]. Two key points are helpful to keep in mind regarding these genetic alterations, with respect to their ability to risk-stratify cytologically indeterminate thyroid nodules.
First, for thyroid FNA-based molecular testing, the detection of mutations and gene fusions is not a binary “positive” or “negative” result. Instead, these tests offer a gradient of cancer risk estimates based on the type of genetic alteration that is detected. Driver alterations, such as the BRAF V600E mutation, and chromosomal rearrangements involving RET, ALK, or NTRK genes are essentially diagnostic of cancer in the context of thyroid tumors [30,34,35]. Detection of one of these alterations typically warrants thyroidectomy for therapeutic purposes, with consideration of total thyroidectomy as the initial surgical approach if the likelihood of requiring adjuvant radioactive iodine treatment is high, based on clinical and sonographic features (e.g., tumor size > 4 cm, suspicion of extrathyroidal extension, lymph node metastasis, or involvement of contralateral lobe). In contrast, mutations in the RAS gene family (HRAS, KRAS, NRAS), BRAF K601E mutation, PAX8-PPARG gene fusion, and gene fusions involving THADA are specific for neoplasia but not necessarily for malignancy; these alterations have been detected in benign (follicular adenoma), pre-malignant (noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP)), and malignant tumors (follicular carcinoma, encapsulated follicular variant of papillary carcinoma) [29,30,36,37,38]. Given their modest specificity for cancer, detection of one of these alterations alone generally warrants diagnostic lobectomy rather than total thyroidectomy as the initial surgical approach.
Second, driver alterations often align with tumor phenotype. The BRAF V600E mutation and other driver alterations that result in similar “BRAF V600E-like” gene expression profiles are associated with classical, tall-cell variant and infiltrative follicular variant papillary carcinomas. In contrast, RAS mutations, BRAF K601E mutation, PAX8-PPARG gene fusion, and gene rearrangements involving THADA are associated with a spectrum of follicular-patterned neoplasms, as indicated above. Mutations in the TP53 and TERT promoter region, particularly in combination with the aforementioned driver alterations, are found with increased frequency in aggressive thyroid cancers, including poorly differentiated carcinoma and anaplastic carcinoma [39,40,41]. Finally, Hürthle cell neoplasms have distinctive mutations in mitochondrial DNA as well as recurrent chromosome-level copy number alterations; additional somatic mutations superimposed on this background, including those involving TERT, TP53, and RAS-family genes, have been reported in Hürthle cell carcinomas [31,32,42,43,44]. In this light, ancillary tests for thyroid FNAs that detect mutations, gene fusions, and copy-number alterations can inform clinicians about a tumor’s phenotype and potentially about its aggressiveness as well. Apart from diagnostic value, the detection of driver alterations in advanced-stage thyroid cancers may identify tumors amenable to targeted therapies [45]. In uncommon cases, genotyping results from a thyroid FNA may also detect genetic alterations associated with familial cancer syndromes, in which case germline genetic testing may be recommended.
2.2. Gene and microRNA Expression Profiles
The different transcriptional profiles of benign and malignant thyroid tumors offer an additional opportunity for risk-stratifying cytologically indeterminate nodules on FNA samples. In contrast to specific driver mutations and gene fusions, gene [46,47,48] and microRNA [49,50,51,52] expression profiles can be understood as readouts of the cumulative and complex genetic, epigenetic, and possibly environmental influences on cells. Large-scale gene expression analysis can currently be performed using high-throughput RNA sequencing technology. For smaller panels, microarray or RT-PCR based tests can be used to detect relative differences in gene or microRNA expression.
3. Molecular Assays for Cytologically Indeterminate Thyroid Nodules
The latest molecular tests for cytologically indeterminate thyroid nodules offer varying degrees of risk stratification, but from a clinical management standpoint, test results can be conceptualized into three broad tiers based on probability of cancer (Figure 1):
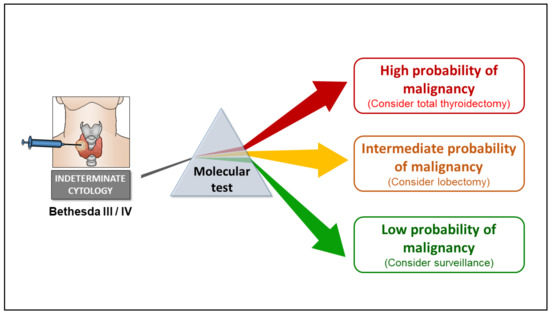
Figure 1.
Model illustrating role of molecular testing for refining the cancer risk stratification of cytologically indeterminate (Bethesda III or IV) thyroid nodules. Current versions of molecular tests described in this review offer varying degrees of risk stratification, but for the most part, test results fall into one of three categories: low, intermediate, or high probability of cancer. A high negative predictive value (NPV) is a key feature shared by all the tests described herein. Among most cohorts of Bethesda III/IV nodules, a NPV greater than 95% (as demonstrated by clinical validation studies) indicates that the cancer risk associated with a negative molecular testing result is less than 5%, similar to that of a cytologically benign nodule.
- (1).
- High-probability results, where the probability of cancer is so high that thyroidectomy is indicated for therapeutic purposes (the decision between lobectomy versus total thyroidectomy may be informed by clinical, sonographic, cytologic, and molecular features of a nodule).
- (2).
- Intermediate-probability results, for which a diagnostic lobectomy is recommended for definitive nodule classification and in many cases may suffice from a therapeutic standpoint.
- (3).
- Low-probability results, where the cancer risk is similar to that of cytologically benign aspirates, for which clinical surveillance would be adequate.
An important feature shared by each of the tests discussed below is a high negative predictive value (NPV), which reflects the test’s ability to recognize the molecular profiles of nodules that are very likely to be benign and can be followed by sonographic surveillance instead of diagnostic surgery.
In the United States, three main molecular tests have been commercially developed over the past decade for cytologically indeterminate thyroid nodules: ThyroSeq v3 (University of Pittsburgh Medical Center and Sonic Healthcare USA, Rye Brook, NY, USA), ThyGeNEXT and ThyraMIR (Interpace Diagnostics, Parsippany, NJ, USA), and Afirma Gene Sequencing Classifier (GSC) and Xpression Atlas (Veracyte, South San Francisco, CA, USA). In their current forms, each of these tests use a combination of genotyping and mRNA or microRNA expression profiling to refine the preoperative cancer risk of cytologically indeterminate thyroid nodules (Table 2).

Table 2.
Comparison of testing methodology, biomarkers, and quality control measures for the three commercially available thyroid molecular testing platforms in the United States.
3.1. ThyroSeq v3
Early versions of molecular tests risk-stratify cytologically indeterminate nodules based on the detection of oncogenic alterations involved a small panel of the most common driver mutations and fusions involving seven genes (BRAF, HRAS, KRAS, NRAS, RET-PTC1, RET-PTC3, PAX8-PPARG). A small genotyping panel stratified nodules into those with high and intermediate probabilities for cancer but was limited by a low negative predictive value (NPV). In reports of institutional experiences with the seven-gene panel, the NPV ranged from 82–94% among Bethesda III/IV aspirates (NIFTP/cancer prevalence of 14–32%); a negative test result was thus associated with a residual cancer risk of 6–18%, making it suboptimal for ruling out malignancy [53,54,55,56].
Over the past decade, the ThyroSeq test has utilized the high throughput of targeted next-generation DNA and RNA sequencing (NGS) platforms as well as data from several large-scale genomic characterizations of thyroid cancers to expand the test panel to 12,135 mutations, insertions, and deletions in 112 genes and over 120 different types of gene fusions. Gene expression alterations and chromosomal copy-number alterations (characteristic of Hürthle cell neoplasms) are also examined using RNA and DNA sequencing, respectively [57].
For thyroid FNAs classified as Bethesda III or IV, nucleic acid for ThyroSeq testing can be extracted from cellular material collected directly into nucleic acid preservative (provided by the vendor), formalin-fixed paraffin-embedded (FFPE) cellblock preparations, and more recently, cells on direct smear slides [58]. ThyroSeq’s limited gene expression analysis serves as quality control steps, confirming adequate thyroid follicular cell sampling as well as identifying aspirates that are not of thyroid follicular cell origin (e.g., medullary thyroid carcinoma (MTC), parathyroid, metastatic tumors). The sequencing results of samples that pass these quality control steps are analyzed by bioinformatics pipelines and assigned a Genomic Classifier (GC) score based on the number, type, and allelic frequency of the genetic alterations that are identified [57]. ThyroSeq currently offers six tiers of test results, which are stratified by probability of cancer or NIFTP, tumor type, and risk of recurrence, which inform their recommendations for patient management.
ThyroSeq v3 was clinically validated in a prospective, multi-institutional, blinded study [59] (Table 3). Both test results and histopathologic classifications were binned into binary outcomes for the purposes of measuring test performance. For ThyroSeq v3 results, the absence of oncogenic alterations and detection of low-risk alterations (particularly those that were detected at low levels by NGS) were pooled together as negative test results, while the remaining alterations covering a spectrum of tumor types, cancer probabilities, and recurrence risk were collectively considered positive test results. For the gold standard diagnoses, positive outcomes consisted of NIFTP as well as low-risk and high-risk cancers (jointly reported as “NIFTP/cancer” in this review, because NIFTP is currently considered a surgical disease for which lobectomy is diagnostically necessary and therapeutically sufficient [60]). The clinical validation study involved 154 Bethesda III and 93 Bethesda IV nodules (combined NIFTP and cancer prevalence of 28%) that were tested with ThyroSeq v3 and reference histopathologic diagnosis. ThyroSeq v3 had a sensitivity of 94% and negative predictive value (NPV) of 97% for NIFTP/cancer in the study population, corresponding to a NIFTP/cancer risk of approximately 3% if no oncogenic alterations were identified. The clinical validation study also reported 82% specificity and 66% positive predictive value (PPV). Importantly, in real-world practice, neither the test results nor histopathologic outcomes are binary. Therefore, PPV as reported in these studies offers a limited view of test performance.

Table 3.
Comparison of clinical validation studies for three commercially available molecular tests for cytologically indeterminate thyroid nodules.
3.2. ThyGeNEXT/ThyraMIR
To improve the NPV of the genotyping approach for refining the risk stratification of cytologically indeterminate nodules, Interpace Diagnostics combines a relatively focused genotyping panel (ThyGeNEXT) with a second test: a microRNA expression classifier optimized to have a high NPV for thyroid cancer. For aspirates classified as Bethesda III or IV, testing can be performed either from cells collected by dedicated FNA passes into a vial of nucleic acid preservative or from cells on direct smear slides that were routinely prepared for cytologic evaluation [61]. Samples first undergo evaluation by ThyGeNEXT, a targeted DNA and RNA NGS panel that currently includes hotspot mutations in 10 genes and 38 gene fusions [62]. Adequate sampling of thyroid follicular cells is determined by a limited gene expression panel that includes NKX2.1 (TTF-1) and PAX8. Detection of driver alterations associated with a high probability of cancer (e.g., BRAF V600E mutation, BRAF fusions, RET fusions, TERT promoter mutations, ALK mutations and fusions)—reported by Interpace Diagnostics as “strong” driver mutations—require no further testing. Samples that are either (a) negative for alterations in the ThyGeNEXT panel or (b) positive for a “weak” driver alteration (e.g., mutations in RAS-family genes) have an intermediate probability of cancer and are risk-stratified further using ThyraMIR.
ThyraMIR uses quantitative RT-PCR to measure the relative expression levels of 10 microRNAs, the results of which are sorted into three risk-based categories. These microRNA expression profiles are combined with the results of the mutation/fusion panel to assign a global estimate of cancer risk for each nodule. Nodules that are negative for both ThyGeNEXT and ThyraMIR are considered to be low risk for NIFTP/cancer, while other combinations of ThyGeNEXT and ThyraMIR results are aligned with a range of NIFTP/cancer risks based on the specific mutations, gene fusions, and microRNA profile that are detected. From a clinical standpoint, the combined test results can be stratified into three tiers (negative, moderate, and positive with respect to cancer probability), warranting consideration of clinical surveillance, lobectomy, or total thyroidectomy, respectively. Medullary carcinoma is recognized by upregulation of specific microRNAs (e.g., miR-375) in the ThyraMIR panel [63].
The combined ThyGeNEXT/ThyraMIR testing approach was clinically validated in a retrospective study of archival, cytologically indeterminate thyroid FNA smears with matched resection specimens that were collected from four institutions [64] (Table 2). Reference histopathologic diagnoses were divided into benign nodules versus those that would currently warrant resection (NIFTP and cancer). Among the 178 Bethesda III and IV nodules in the study, the prevalence of NIFTP and cancer was 30%. This clinical validation study diverges from those of ThyroSeq v3 and Afirma GSC in two important ways. First, the ThyroSeq/ThyraMIR validation study takes a unique approach to address the limitations of using a conventional 2 × 2 matrix for assessing diagnostic accuracy of non-binary classification tests. Although ThyGeNEXT/ThyraMIR results are binned into three tiers as described above, the middle tier (moderate-risk) test results are alternatively considered “positive” or “negative”, depending on the statistical measure being calculated: moderate-risk results are grouped together with positive test results for calculating test sensitivity and NPV, while the same moderate-risk results are grouped with negative test results for calculating test specificity and PPV. Second, the authors also noted that the prevalence of NIFTP and cancer among samples selected for their validation cohort (30%) was higher than what was found in prior studies [65]. Bayes’ theorem was thus applied to extrapolate the expected rate of negative, moderate, and positive test results at a lower (14%) NIFTP/cancer prevalence. Altogether, using these two adjustments, Lupo et al. report 95% sensitivity, 90% specificity, 97% NPV, and 75% PPV for ThyGeNEXT/ThyraMIR. For the purposes of comparison with the other commercially available molecular tests described herein, we also provide corresponding values for test performance that were calculated using conventional methods (i.e., considering moderate-risk test results as “positive”; no prevalence adjustment): 93% sensitivity, 62% specificity, 95% NPV, and 52% PPV. Importantly, with both approaches, cytologically indeterminate aspirates that are negative for both ThyGeNEXT and ThyraMIR have a low cancer risk (3–5%) based on 95–97% NPV of the combined test.
3.3. Afirma GSC and Xpression Atlas
Afirma refines the cancer risk of cytologically indeterminate thyroid nodules by evaluating the expression profile of a large panel of genes and using machine-learning algorithms to classify aspirates as having either a “Benign” or “Suspicious” gene expression profile. The initial version of the test, introduced in 2012 as the Afirma Gene Expression Classifier (GEC), relied on expression microarrays to determine the expression profile of 142 genes [66]. Over the past decade, Afirma has upgraded their microarray-based GEC to a high-throughput RNA sequencing platform that determines the expression profile of 10,196 genes. Updated machine-learning algorithms were applied to develop the current Gene Sequencing Classifier (GSC), which risk-stratifies aspirates based on the expression pattern of 1115 core genes; the remaining ~9000 genes, whose expression levels are measured, reportedly provide stability to the classifier model. The GSC has also added mitochondrial transcripts and loss-of-heterozygosity analysis to its test panel in an effort to improve risk stratification of Hürthle cell lesions [67].
Afirma GSC requires dedicated FNA passes to be collected into a vial of nucleic acid preservative, in addition to the cellular material collected for routine cytological evaluation. For aspirates classified as Bethesda III or IV, the concurrent material collected for molecular testing is processed for RNA sequencing. Expression profiles that indicate sampling of parathyroid tissue or medullary carcinoma (Afirma MTC test) are reported as such and require no further testing. Similarly, the expression profile associated with BRAF V600E mutation (Afirma BRAF test) as well as detection of RET-PTC1 or RET-PTC3 gene fusions are highly specific for papillary carcinoma and do not require further analysis by the GSC. For the remaining samples that are (a) negative for the above markers and (b) confirmed to have adequate thyroid follicular cell content, the GSC’s proprietary algorithm classifies each sample as having either a “Benign” or a “Suspicious” expression profile.
The original version of the Afirma test (GEC) was clinically validated in a multicenter prospective study involving 210 Bethesda III and IV aspirates with matched resection specimens [66]. Archival patient samples remaining from this original GEC validation study were used to validate the current version of Afirma (GSC); 190 Bethesda III/IV samples had sufficient RNA available for the GSC validation, with similar prevalence of NIFTP/cancer (24%) to the original 210-sample GEC validation set [68] (Table 2). The GSC recognized 54% of Bethesda III/IV nodules in the validation set as having a “Benign” gene expression profile, compared to the 41% benign-call rate of the GEC. Compared to the GEC (90% sensitivity, 52% specificity, 94% NPV, and 37% PPV), the GSC showed better specificity (68%) and PPV (47%) for NIFTP/cancer, while maintaining the high sensitivity (91%) and NPV (96%) of the original test. Altogether, a Benign GSC result is thus associated with an approximate 4% risk of NIFTP/cancer (corresponding to 1-NPV), while a Suspicious GSC result confers a NIFTP/cancer risk of approximately 47% (corresponding to the PPV). Among the 26 Hürthle cell lesions (9 Hürthle cell carcinomas and 17 Hürthle cell adenomas) in the validation set, the GSC showed improved specificity (59%), NPV (91%), and PPV (53%) compared to the GEC (12% specificity, 67% NPV, and 35% PPV).
While measurement of gene expression levels by RNA sequencing is the core methodology for the GSC, the sequencing data can also be mined for point mutations, insertions/deletions, and gene fusions. Sequence variants that are known to be associated with thyroid cancer are reported in a complementary test known as Afirma Xpression Atlas [69]. As an RNA sequencing-based test, Xpression Atlas currently reports 235 fusions and 905 sequence variants from only the transcribed portion of 593 genes [70]. Alterations in non-coding DNA, such as those in the TERT promoter region, are not detected by the Xpression Atlas. Like ThyroSeq and ThyGeNEXT, the Afirma Xpression Atlas provides patients and clinicians with estimates of NIFTP/cancer risk and tumor type based on the specific variant or fusion that is identified. As discussed above for genotyping panels, the detection of oncogenic mutations and gene fusions has potential benefits beyond diagnostic utility. Identification of actionable driver alterations may guide the selection of targeted therapy for patients with advanced thyroid cancers, and detection of alterations suggestive of heritable cancer syndromes may steer patients towards genetic counseling and testing for germline alterations.
4. Conclusions
While ThyroSeq v3, ThyGeNEXT/ThyraMIR, and Afirma GSC/Xpression Atlas each take different approaches to refine the cancer risk stratification of thyroid nodules, common themes have emerged as these tests have evolved over the years. A high NPV to help rule-out malignancy is key among these similarities, as are combined testing approaches that use aspects of both genotyping and gene or microRNA expression profiling to offer more granular estimates of tumor phenotype and cancer risk. Ultimately, each of these three testing platforms appear to provide similar information to the patient and clinician in terms of guiding management decisions. The molecular tests described in this review have primarily been validated and used in the United States. International differences in the application and impact of molecular testing for thyroid FNA samples have been reported and likely reflect contrasting management strategies for cytologically indeterminate nodules as well as differences in access to testing among different countries [25]. In our increasingly interconnected world, we expect that global discussions about our diverse risk-stratification approaches for thyroid nodules will help us move towards safer and more cost-effective care for patients worldwide.
Author Contributions
Conceptualization, M.N., C.B. and Z.B.; writing—original draft preparation, M.N. and Z.B.; writing—review and editing, M.N., C.B. and Z.B. All authors have read and agreed to the published version of this manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Pacini, F.; Schlumberger, M.; et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid 2009, 19, 1167–1214. [Google Scholar] [CrossRef]
- Hirokawa, M.; Suzuki, A.; Higuchi, M.; Hayashi, T.; Kuma, S.; Ito, Y.; Miyauchi, A. The Japanese reporting system for thyroid aspiration cytology 2019 (JRSTAC2019). Gland Surg. 2020, 9, 1653–1662. [Google Scholar] [CrossRef]
- Vuong, H.G.; Ngo, H.T.T.; Bychkov, A.; Jung, C.K.; Vu, T.H.; Lu, K.B.; Kakudo, K.; Kondo, T. Differences in surgical resection rate and risk of malignancy in thyroid cytopathology practice between Western and Asian countries: A systematic review and meta-analysis. Cancer Cytopathol. 2020, 128, 238–249. [Google Scholar] [CrossRef]
- Davies, L.; Morris, L.G.; Haymart, M.; Chen, A.Y.; Goldenberg, D.; Morris, J.; Ogilvie, J.B.; Terris, D.J.; Netterville, J.; Wong, R.J.; et al. American Association of Clinical Endocrinologists and American College of Endocrinology Disease State Clinical Review: The Increasing Incidence of Thyroid Cancer. Endocr. Pract. 2015, 21, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Frates, M.C.; Benson, C.B.; Charboneau, J.W.; Cibas, E.S.; Clark, O.H.; Coleman, B.G.; Cronan, J.J.; Doubilet, P.M.; Evans, D.B.; Goellner, J.R.; et al. Management of thyroid nodules detected at US: Society of Radiologists in Ultrasound consensus conference statement. Ultrasound Q. 2006, 22, 231–238; discussion 239–240. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.H.; Suh, C.H.; Baek, J.H.; Chung, S.R.; Choi, Y.J.; Lee, J.H. Unnecessary thyroid nodule biopsy rates under four ultrasound risk stratification systems: A systematic review and meta-analysis. Eur. Radiol. 2021, 31, 2877–2885. [Google Scholar] [CrossRef] [PubMed]
- Trimboli, P.; Knappe, L.; Treglia, G.; Ruberto, T.; Piccardo, A.; Ceriani, L.; Paone, G.; Giovanella, L. FNA indication according to ACR-TIRADS, EU-TIRADS and K-TIRADS in thyroid incidentalomas at (18)F-FDG PET/CT. J. Endocrinol. Investig. 2020, 43, 1607–1612. [Google Scholar] [CrossRef]
- Mistry, R.; Hillyar, C.; Nibber, A.; Sooriyamoorthy, T.; Kumar, N. Ultrasound Classification of Thyroid Nodules: A Systematic Review. Cureus 2020, 12, e7239. [Google Scholar] [CrossRef]
- Castellana, M.; Castellana, C.; Treglia, G.; Giorgino, F.; Giovanella, L.; Russ, G.; Trimboli, P. Performance of Five Ultrasound Risk Stratification Systems in Selecting Thyroid Nodules for FNA. J. Clin. Endocrinol. Metab. 2020, 105. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.Z.; Cibas, E.S. The Bethesda System for Reporting Thyroid Cytopathology: Definitions, Criteria, and Explanatory Notes; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Trimboli, P.; Crescenzi, A.; Castellana, M.; Giorgino, F.; Giovanella, L.; Bongiovanni, M. Italian consensus for the classification and reporting of thyroid cytology: The risk of malignancy between indeterminate lesions at low or high risk. A systematic review and meta-analysis. Endocrine 2019, 63, 430–438. [Google Scholar] [CrossRef]
- Poller, D.N.; Bongiovanni, M.; Trimboli, P. Risk of malignancy in the various categories of the UK Royal College of Pathologists Thy terminology for thyroid FNA cytology: A systematic review and meta-analysis. Cancer Cytopathol. 2020, 128, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Poller, D.N.; Baloch, Z.W.; Fadda, G.; Johnson, S.J.; Bongiovanni, M.; Pontecorvi, A.; Cochand-Priollet, B. Thyroid FNA: New classifications and new interpretations. Cancer Cytopathol. 2016, 124, 457–466. [Google Scholar] [CrossRef]
- Poller, D.N.; Schmitt, F. Should uncertainty concerning the risk of malignancy be included in diagnostic (nongynecologic) cytopathology reports? Cancer Cytopathol. 2021, 129, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.P.X.; Truong, V.T.; Kakudo, K.; Vuong, H.G. The diversities in thyroid cytopathology practices among Asian countries using the Bethesda system for reporting thyroid cytopathology. Gland Surg. 2020, 9, 1735–1746. [Google Scholar] [CrossRef]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Sherman, S.I.; Tuttle, R.M. Management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid 2006, 16, 109–142. [Google Scholar] [CrossRef] [PubMed]
- Nikiforov, Y.E.; Seethala, R.R.; Tallini, G.; Baloch, Z.W.; Basolo, F.; Thompson, L.D.; Barletta, J.A.; Wenig, B.M.; Al Ghuzlan, A.; Kakudo, K.; et al. Nomenclature Revision for Encapsulated Follicular Variant of Papillary Thyroid Carcinoma: A Paradigm Shift to Reduce Overtreatment of Indolent Tumors. JAMA Oncol. 2016, 2, 1023–1029. [Google Scholar] [CrossRef]
- Baloch, Z.W.; Seethala, R.R.; Faquin, W.C.; Papotti, M.G.; Basolo, F.; Fadda, G.; Randolph, G.W.; Hodak, S.P.; Nikiforov, Y.E.; Mandel, S.J. Noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP): A changing paradigm in thyroid surgical pathology and implications for thyroid cytopathology. Cancer Cytopathol. 2016, 124, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.V.; Osamura, R.Y.; Kloppel, G.; Rosai, J. (Eds.) WHO Classification of Tumours of Endocrine Organs, 4th ed.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; Volume 10.
- Nikiforov, Y.E.; Baloch, Z.W.; Hodak, S.P.; Giordano, T.J.; Lloyd, R.V.; Seethala, R.R.; Wenig, B.M. Change in Diagnostic Criteria for Noninvasive Follicular Thyroid Neoplasm with Papillarylike Nuclear Features. JAMA Oncol. 2018, 4, 1125–1126. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.D.; Faquin, W.C.; Baloch, Z.; Fadda, G.; Thompson, L.; Larocca, L.M.; Pantanowitz, L. Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features (NIFTP): Update and Diagnostic Considerations—A Review. Endocr. Pathol. 2019, 30, 155–162. [Google Scholar] [CrossRef]
- Zhou, H.; Baloch, Z.W.; Nayar, R.; Bizzarro, T.; Fadda, G.; Adhikari-Guragain, D.; Hatem, J.; Larocca, L.M.; Samolczyk, J.; Slade, J.; et al. Noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP): Implications for the risk of malignancy (ROM) in the Bethesda System for Reporting Thyroid Cytopathology (TBSRTC). Cancer Cytopathol. 2018, 126, 20–26. [Google Scholar] [CrossRef]
- Nishino, M.; Nikiforova, M. Update on Molecular Testing for Cytologically Indeterminate Thyroid Nodules. Arch. Pathol. Lab. Med. 2018, 142, 446–457. [Google Scholar] [CrossRef]
- Ngo, H.T.T.; Nguyen, T.P.X.; Vu, T.H.; Jung, C.K.; Hassell, L.; Kakudo, K.; Vuong, H.G. Impact of Molecular Testing on the Management of Indeterminate Thyroid Nodules Among Western and Asian Countries: A Systematic Review and Meta-analysis. Endocr. Pathol. 2020. [Google Scholar] [CrossRef]
- Ohori, N.P.; Landau, M.S.; Manroa, P.; Schoedel, K.E.; Seethala, R.R. Molecular-derived estimation of risk of malignancy for indeterminate thyroid cytology diagnoses. J. Am. Soc. Cytopathol. 2020, 9, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Nylen, C.; Mechera, R.; Marechal-Ross, I.; Tsang, V.; Chou, A.; Gill, A.J.; Clifton-Bligh, R.J.; Robinson, B.G.; Sywak, M.S.; Sidhu, S.B.; et al. Molecular Markers Guiding Thyroid Cancer Management. Cancers 2020, 12, 2164. [Google Scholar] [CrossRef]
- Kumar, N.; Gupta, R.; Gupta, S. Molecular testing in diagnosis of indeterminate thyroid cytology: Trends and drivers. Diagn. Cytopathol. 2020, 48, 1144–1151. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Lee, S.; Kim, S.J.; Jee, H.G.; Kim, B.A.; Cho, H.; Song, Y.S.; Cho, S.W.; Won, J.K.; Shin, J.Y.; et al. Comprehensive Analysis of the Transcriptional and Mutational Landscape of Follicular and Papillary Thyroid Cancers. PLoS Genet. 2016, 12, e1006239. [Google Scholar] [CrossRef]
- Ganly, I.; Makarov, V.; Deraje, S.; Dong, Y.; Reznik, E.; Seshan, V.; Nanjangud, G.; Eng, S.; Bose, P.; Kuo, F. Integrated genomic analysis of Hürthle Cell cancer reveals oncogenic drivers, recurrent mitochondrial mutations, and unique chromosomal landscapes. Cancer Cell 2018, 34, 256–270.e255. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.K.; Kübler, K.; Calvo, S.E.; Polak, P.; Livitz, D.; Rosebrock, D.; Sadow, P.M.; Campbell, B.; Donovan, S.E.; Amin, S. Widespread chromosomal losses and mitochondrial DNA alterations as genetic drivers in Hürthle cell carcinoma. Cancer Cell 2018, 34, 242–255.e245. [Google Scholar] [CrossRef]
- Giordano, T.J. Genomic hallmarks of thyroid neoplasia. Annu. Rev. Pathol. Mech. Dis. 2018, 13, 141–162. [Google Scholar] [CrossRef]
- Chu, Y.H.; Dias-Santagata, D.; Farahani, A.A.; Boyraz, B.; Faquin, W.C.; Nose, V.; Sadow, P.M. Clinicopathologic and molecular characterization of NTRK-rearranged thyroid carcinoma (NRTC). Mod. Pathol. 2020, 33, 2186–2197. [Google Scholar] [CrossRef]
- Panebianco, F.; Nikitski, A.V.; Nikiforova, M.N.; Kaya, C.; Yip, L.; Condello, V.; Wald, A.I.; Nikiforov, Y.E.; Chiosea, S.I. Characterization of thyroid cancer driven by known and novel ALK fusions. Endocr. Relat. Cancer 2019, 26, 803–814. [Google Scholar] [CrossRef]
- McFadden, D.G.; Dias-Santagata, D.; Sadow, P.M.; Lynch, K.D.; Lubitz, C.; Donovan, S.E.; Zheng, Z.; Le, L.; Iafrate, A.J.; Daniels, G.H. Identification of oncogenic mutations and gene fusions in the follicular variant of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2014, 99, E2457–E2462. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, D.W.; Heo, Y.J.; Baek, J.W.; Lee, Y.J.; Choo, H.J.; Park, Y.M.; Park, H.K.; Ha, T.K.; Kim, D.H.; et al. Utility of including BRAF mutation analysis with ultrasonographic and cytological diagnoses in ultrasonography-guided fine-needle aspiration of thyroid nodules. PLoS ONE 2018, 13, e0202687. [Google Scholar] [CrossRef]
- Najafian, A.; Noureldine, S.; Azar, F.; Atallah, C.; Trinh, G.; Schneider, E.B.; Tufano, R.P.; Zeiger, M.A. RAS Mutations, and RET/PTC and PAX8/PPAR-gamma Chromosomal Rearrangements Are Also Prevalent in Benign Thyroid Lesions: Implications Thereof and A Systematic Review. Thyroid 2017, 27, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Sykorova, V.; Dvorakova, S.; Vcelak, J.; Vaclavikova, E.; Halkova, T.; Kodetova, D.; Lastuvka, P.; Betka, J.; Vlcek, P.; Reboun, M.; et al. Search for new genetic biomarkers in poorly differentiated and anaplastic thyroid carcinomas using next generation sequencing. Anticancer Res. 2015, 35, 2029–2036. [Google Scholar]
- Kunstman, J.W.; Juhlin, C.C.; Goh, G.; Brown, T.C.; Stenman, A.; Healy, J.M.; Rubinstein, J.C.; Choi, M.; Kiss, N.; Nelson-Williams, C.; et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum. Mol. Genet. 2015, 24, 2318–2329. [Google Scholar] [CrossRef]
- Xu, B.; Ghossein, R. Genomic Landscape of Poorly Differentiated and Anaplastic Thyroid Carcinoma. Endocr. Pathol. 2016, 27, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Corver, W.E.; Ruano, D.; Weijers, K.; den Hartog, W.C.; van Nieuwenhuizen, M.P.; de Miranda, N.; van Eijk, R.; Middeldorp, A.; Jordanova, E.S.; Oosting, J.; et al. Genome haploidisation with chromosome 7 retention in oncocytic follicular thyroid carcinoma. PLoS ONE 2012, 7, e38287. [Google Scholar] [CrossRef] [PubMed]
- Kasaian, K.; Chindris, A.M.; Wiseman, S.M.; Mungall, K.L.; Zeng, T.; Tse, K.; Schein, J.E.; Rivera, M.; Necela, B.M.; Kachergus, J.M.; et al. MEN1 mutations in Hurthle cell (oncocytic) thyroid carcinoma. J. Clin. Endocrinol. Metab. 2015, 100, E611–E615. [Google Scholar] [CrossRef]
- Ganly, I.; McFadden, D.G. Short Review: Genomic Alterations in Hurthle Cell Carcinoma. Thyroid 2019, 29, 471–479. [Google Scholar] [CrossRef]
- Lubitz, C.C.; Sadow, P.M.; Daniels, G.H.; Wirth, L.J. Progress in Treating Advanced Thyroid Cancers in the Era of Targeted Therapy. Thyroid 2021. [Google Scholar] [CrossRef]
- Chudova, D.; Wilde, J.I.; Wang, E.T.; Wang, H.; Rabbee, N.; Egidio, C.M.; Reynolds, J.; Tom, E.; Pagan, M.; Rigl, C.T.; et al. Molecular classification of thyroid nodules using high-dimensionality genomic data. J. Clin. Endocrinol. Metab. 2010, 95, 5296–5304. [Google Scholar] [CrossRef]
- Eszlinger, M.; Krohn, K.; Frenzel, R.; Kropf, S.; Tonjes, A.; Paschke, R. Gene expression analysis reveals evidence for inactivation of the TGF-beta signaling cascade in autonomously functioning thyroid nodules. Oncogene 2004, 23, 795–804. [Google Scholar] [CrossRef]
- Eszlinger, M.; Wiench, M.; Jarzab, B.; Krohn, K.; Beck, M.; Lauter, J.; Gubala, E.; Fujarewicz, K.; Swierniak, A.; Paschke, R. Meta- and reanalysis of gene expression profiles of hot and cold thyroid nodules and papillary thyroid carcinoma for gene groups. J. Clin. Endocrinol. Metab. 2006, 91, 1934–1942. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rossi, E.D.; Bizzarro, T.; Martini, M.; Capodimonti, S.; Sarti, D.; Cenci, T.; Bilotta, M.; Fadda, G.; Larocca, L.M. The evaluation of miRNAs on thyroid FNAC: The promising role of miR-375 in follicular neoplasms. Endocrine 2016, 54, 723–732. [Google Scholar] [CrossRef]
- Dettmer, M.; Vogetseder, A.; Durso, M.B.; Moch, H.; Komminoth, P.; Perren, A.; Nikiforov, Y.E.; Nikiforova, M.N. MicroRNA expression array identifies novel diagnostic markers for conventional and oncocytic follicular thyroid carcinomas. J. Clin. Endocrinol. Metab. 2013, 98, E1–E7. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Yip, L.; Kelly, L.; Shuai, Y.; Armstrong, M.J.; Nikiforov, Y.E.; Carty, S.E.; Nikiforova, M.N. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann. Surg. Oncol. 2011, 18, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Labourier, E.; Shifrin, A.; Busseniers, A.E.; Lupo, M.A.; Manganelli, M.L.; Andruss, B.; Wylie, D.; Beaudenon-Huibregtse, S. Molecular Testing for miRNA, mRNA, and DNA on Fine-Needle Aspiration Improves the Preoperative Diagnosis of Thyroid Nodules With Indeterminate Cytology. J. Clin. Endocrinol. Metab. 2015, 100, 2743–2750. [Google Scholar] [CrossRef] [PubMed]
- Eszlinger, M.; Krogdahl, A.; Munz, S.; Rehfeld, C.; Precht Jensen, E.M.; Ferraz, C.; Bosenberg, E.; Drieschner, N.; Scholz, M.; Hegedus, L.; et al. Impact of molecular screening for point mutations and rearrangements in routine air-dried fine-needle aspiration samples of thyroid nodules. Thyroid 2014, 24, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Eszlinger, M.; Piana, S.; Moll, A.; Bosenberg, E.; Bisagni, A.; Ciarrocchi, A.; Ragazzi, M.; Paschke, R. Molecular testing of thyroid fine-needle aspirations improves presurgical diagnosis and supports the histologic identification of minimally invasive follicular thyroid carcinomas. Thyroid 2015, 25, 401–409. [Google Scholar] [CrossRef]
- Nikiforov, Y.E.; Ohori, N.P.; Hodak, S.P.; Carty, S.E.; LeBeau, S.O.; Ferris, R.L.; Yip, L.; Seethala, R.R.; Tublin, M.E.; Stang, M.T.; et al. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: A prospective analysis of 1056 FNA samples. J. Clin. Endocrinol. Metab. 2011, 96, 3390–3397. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Mercurio, S.; Wald, A.I.; Barbi de Moura, M.; Callenberg, K.; Santana-Santos, L.; Gooding, W.E.; Yip, L.; Ferris, R.L.; Nikiforov, Y.E. Analytical performance of the ThyroSeq v3 genomic classifier for cancer diagnosis in thyroid nodules. Cancer 2018, 124, 1682–1690. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Lepe, M.; Tolino, L.A.; Miller, M.E.; Ohori, N.P.; Wald, A.I.; Landau, M.S.; Kaya, C.; Malapelle, U.; Bellevicine, C.; et al. Thyroid cytology smear slides: An untapped resource for ThyroSeq testing. Cancer Cytopathol. 2021, 129, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Steward, D.L.; Carty, S.E.; Sippel, R.S.; Yang, S.P.; Sosa, J.A.; Sipos, J.A.; Figge, J.J.; Mandel, S.; Haugen, B.R.; Burman, K.D.; et al. Performance of a Multigene Genomic Classifier in Thyroid Nodules With Indeterminate Cytology: A Prospective Blinded Multicenter Study. JAMA Oncol. 2019, 5, 204–212. [Google Scholar] [CrossRef]
- Haugen, B.R.; Sawka, A.M.; Alexander, E.K.; Bible, K.C.; Caturegli, P.; Doherty, G.M.; Mandel, S.J.; Morris, J.C.; Nassar, A.; Pacini, F.; et al. American Thyroid Association Guidelines on the Management of Thyroid Nodules and Differentiated Thyroid Cancer Task Force Review and Recommendation on the Proposed Renaming of Encapsulated Follicular Variant Papillary Thyroid Carcinoma Without Invasion to Noninvasive Follicular Thyroid Neoplasm with Papillary-Like Nuclear Features. Thyroid 2017, 27, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Timmaraju, V.A.; Song-Yang, J.W.; Repko, B.; Narick, C.; Mireskandari, A.; Finkelstein, S. Utility of microdissected cytology smears for molecular analysis of thyroid malignancy. Diagn. Cytopathol. 2019, 47, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Ablordeppey, K.K.; Timmaraju, V.A.; Song-Yang, J.W.; Yaqoob, S.; Narick, C.; Mireskandari, A.; Finkelstein, S.D.; Kumar, G. Development and Analytical Validation of an Expanded Mutation Detection Panel for Next-Generation Sequencing of Thyroid Nodule Aspirates. J. Mol. Diagn. 2020, 22, 355–367. [Google Scholar] [CrossRef]
- Ciarletto, A.M.; Narick, C.; Malchoff, C.D.; Massoll, N.A.; Labourier, E.; Haugh, K.; Mireskandari, A.; Finkelstein, S.D.; Kumar, G. Analytical and clinical validation of pairwise microRNA expression analysis to identify medullary thyroid cancer in thyroid fine-needle aspiration samples. Cancer Cytopathol. 2021, 129, 239–249. [Google Scholar] [CrossRef]
- Lupo, M.A.; Walts, A.E.; Sistrunk, J.W.; Giordano, T.J.; Sadow, P.M.; Massoll, N.; Campbell, R.; Jackson, S.A.; Toney, N.; Narick, C.M.; et al. Multiplatform molecular test performance in indeterminate thyroid nodules. Diagn. Cytopathol. 2020, 48, 1254–1264. [Google Scholar] [CrossRef]
- Sistrunk, J.W.; Shifrin, A.; Frager, M.; Bardales, R.H.; Thomas, J.; Fishman, N.; Goldberg, P.; Guttler, R.; Grant, E. Clinical impact of testing for mutations and microRNAs in thyroid nodules. Diagn. Cytopathol. 2019, 47, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Alexander, E.K.; Kennedy, G.C.; Baloch, Z.W.; Cibas, E.S.; Chudova, D.; Diggans, J.; Friedman, L.; Kloos, R.T.; LiVolsi, V.A.; Mandel, S.J.; et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N. Engl. J. Med. 2012, 367, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Duh, Q.Y.; Kloos, R.T.; Babiarz, J.; Harrell, R.M.; Traweek, S.T.; Kim, S.Y.; Fedorowicz, G.; Walsh, P.S.; Sadow, P.M.; et al. Identification of Hurthle cell cancers: Solving a clinical challenge with genomic sequencing and a trio of machine learning algorithms. BMC Syst. Biol. 2019, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.N.; Angell, T.E.; Babiarz, J.; Barth, N.M.; Blevins, T.; Duh, Q.Y.; Ghossein, R.A.; Harrell, R.M.; Huang, J.; Kennedy, G.C.; et al. Performance of a Genomic Sequencing Classifier for the Preoperative Diagnosis of Cytologically Indeterminate Thyroid Nodules. JAMA Surg. 2018, 153, 817–824. [Google Scholar] [CrossRef]
- Angell, T.E.; Wirth, L.J.; Cabanillas, M.E.; Shindo, M.L.; Cibas, E.S.; Babiarz, J.E.; Hao, Y.; Kim, S.Y.; Walsh, P.S.; Huang, J.; et al. Analytical and Clinical Validation of Expressed Variants and Fusions from the Whole Transcriptome of Thyroid FNA Samples. Front. Endocrinol. 2019, 10, 612. [Google Scholar] [CrossRef]
- Krane, J.F.; Cibas, E.S.; Endo, M.; Marqusee, E.; Hu, M.I.; Nasr, C.E.; Waguespack, S.G.; Wirth, L.J.; Kloos, R.T. The Afirma Xpression Atlas for thyroid nodules and thyroid cancer metastases: Insights to inform clinical decision-making from a fine-needle aspiration sample. Cancer Cytopathol. 2020, 128, 452–459. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).