Abstract
(1) Background: Human papillomaviruses (HPVs) are known to be related to the development of about 5% of all human cancers. The clinical relevance of HPV infection has been deeply investigated in carcinomas of the oropharyngeal area, uterine cervix, and anogenital area. To date, several different methods have been used for detecting HPV infection. The aim of the present study was to compare three different methods for the diagnosis of the presence of the HPV genome. (2) Methods: A total of 50 samples were analyzed. Twenty-five of them were tested using both next generation sequencing (NGS) and VisionArray® technology, the other 25 were tested using Hybrid Capture (HC) II assay and VisionArray® technology. (3) Results: A substantial agreement was obtained using NGS and VisionArray® (κ = 0.802), as well as between HC II and VisionArray® (κ = 0.606). In both analyses, the concordance increased if only high risk HPVs I(HR-HPVs) were considered as “positive”. (4) Conclusions: Our data highlighted the importance of technical choice in HPV characterization, which should be guided by the clinical aims, costs, starting material, and turnaround time for results.
1. Introduction
Human papillomaviruses (HPVs) represent a group of more than 150 double-stranded circular DNA viruses involved in the infection of basal cells of the stratified epithelium [1]. HPVs are known to be related to the development of about 5% of all human cancers [2,3,4], and they can be divided into three subtypes based on the clinical prognosis of their associated lesions: (i) high-risk types (HR-HPVs) are known to cause lesions that often have a malignant progression; (ii) probably high-risk types (PHR-HPV); (iii) low-risk types (LR-HPV) are involved in benign epithelial hyperplasias such as warts and recurrent respiratory papillomas [5,6].
Twelve different HPV types are considered to have similar high-risk behavior (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59) [7], though the number of putative HR-HPVs varies from 13 to 19 [8,9,10,11]. HR-HPVs are known to be involved in a subset of carcinomas, such as squamous cell carcinomas (SCCs) in the oropharyngeal area, uterine cervix, and anogenital area. The vast majority (~84%) of all HPV-related cancers are represented by cervical cancer [12]. In the anogenital region, the infection is sexually transmitted and it shows a peak prevalence in women younger than 25 years [6,13]; in the oropharyngeal region, HPV-positive SCCs typically occur in younger non-smoking patients, and they are associated with improved overall outcomes with respect to HPV-negative cancers [14,15].
To date, several different methods are used for detecting HPV infection. The DNA multiplex tandem polymerase chain reaction, RNA or in situ hybridization (ISH), and expression of p16 can be combined for the determination of virus presence in different specimens. Since the publication in the literature of MY and GP primers targeted to the L1 region of the HPV genome [16,17,18], PCR-based methods and sequencing have been considered the “reference standard” [19].
Different diagnostic methods mean different sensitivity and specificity. Molecular approaches have shown high sensitivity when compared to ISH, but they carry the risk of “false positive” (HPV non-cancer-associated) results.
There are many tests that are FDA approved for IVD (in vitro diagnostic) diagnosis, such as Anyplex™ II HPV28 Detection (SeeGene Inc., Seoul, Korea), Cobas® HPV (Roche Molecular Systems, Inc., Branchburg, NJ, USA), HPV OncoTect® E6, E7 mRNA assay (incellDx, San Carlos, CA, USA), Cervista™ HPV HR (Hologic, Ltd., Manchester, UK), Hybrid Capture® 2 High-Risk HPV DNA Test™ (Qiagen, Gaithersburg, MD, USA), and VisionArray® HPV Chips assay (ZytoVision GmbH, Bremerhaven, Germany). Except for VisionArray® technology, which also identifies LR-HPV, PHR-HPV, and HR-HPV, the other assays are targeted only to HR-HPV diagnosis.
The Hybrid Capture (HC) II High-Risk HPV DNA Test allows for the detection of the presence of 13 high-risk HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) in cervical specimens, using RNA probes complementary to HPV DNA, specific antibodies to identify RNA–DNA hybrids and amplify signal amplification, and chemiluminescent detection. Specimens containing the target DNA hybridize with a specific HPV RNA probe.
The HPV test using next generation sequencing (NGS) is a PCR- based analysis that allows for the identification of both HR- and LR-HPV sequencing of the HPV genome and for aligning the results with sequences registered in an HPV database. NGS is a high-throughput sequencing technique that has been widely used to determine the presence of HPV in human specimens using large or small-targeted panels [20,21,22,23,24,25,26,27,28,29,30,31,32,33]. The NGS panel used for HPV detection may be performed with or without a prior PCR. This PCR step may lead to a higher sensitivity, but it could also lead to false-negative results due to eventual mismatches in the sequences of the primers used [22,25,33].
VisionArray® HPV Chips (ZytoVision GmbH, Bremerhaven, Germany) assay is based on DNA amplification by PCR and uses the VisionArray® HPV Primer Kit (ZytoVision GmbH, Bremerhaven, Germany) that is targeted to the L1 region of HPV genomes and allows for the identification of the following HPV types: (i) LR-HPV (6, 11, 40, 42, 43, 44, 54, 55, 57, 61, 62, 72, 81CP8304, 83MM7, 84MM8, 90, and 91); (ii) PHR-HPV (26, 34, 53, 66, 67, 68a, 68b, 69, 70, 73, 82IS39, and 82MM4); (iii) HR-HPV (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59). HPV type is then detected using hybridization between the amplified sequences and the complementary DNA probe, which is captured on a glass chip and analyzed using the VisionArray® Analyzer Software (ZytoVision GmbH, Bremerhaven, Germany).
The present study aimed to compare three different methods (HC II, next generation sequencing, and VisionArray® assay) for the diagnosis of the presence of the HPV genome starting from both formalin-fixed and paraffin-embedded (FFPE) and “fresh-frozen” specimens.
This comparison analyzed the pros and cons of each tested method to help to understand which could be the more appropriate technique according to the clinical aims, available specimen material, the costs, and turnaround time.
2. Methods and Materials
2.1. Sample Collection
By comparing the frequencies of HPV in oropharyngeal and anogenital lesions and those reported in the database of our institution, we planned to analyze a total of 50 samples randomly selected from the archive of Molecular Pathology Laboratory (Bologna). The 50 samples were 11 (22%) specimens from oropharyngeal sites and 39 (78%) from the anogenital region (Table 1). The material analyzed was from 13 FFPE specimens and 37 “fresh-frozen” ones (cervical cytology brushing or non “formalin-fixed and paraffin-embedded” biopsies) (Table 1). The age of the patients ranged from 19 to 70 years old (median 45.0 years). Twelve of the 50 subjects (24.0%) were male and 38 (76.0%) were female.

Table 1.
Cases analyzed for human papillomavirus (HPV) infection.
2.2. DNA Extraction
FFPE samples: DNA was extracted from two to four 10 µm-thick sections using the QuickExtract FFPE DNA Extraction Kit (Epicentre, Madison, WI, USA) by scraping the area of interest according to the selection by a pathologist on the final Hematoxylin and Eosin (H&E) section.
“Fresh-frozen” specimens: DNA was extracted using the MasterPure DNA Purification Kit (Epicentre, Madison, WI, USA), according to the manufacturer’s instruction.
DNA was quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The DNA concentration ranged from 2 ng/μL to 870 ng/μL.
2.3. Next Generation Sequencing
A total of 25 samples (S1–S25, Table 1) were analyzed using the NGS 454 GS-Junior (Roche Diagnostic, Mannheim, Germany) (Figure 1). Thirteen of the 25 (52%) were FFPE specimens and 12 (48%) were “non-fixed” samples (brushing or biopsy). Eleven of the 25 samples (44%) were from the oropharyngeal region and 14 (56%) from the anogenital region (Table 1).
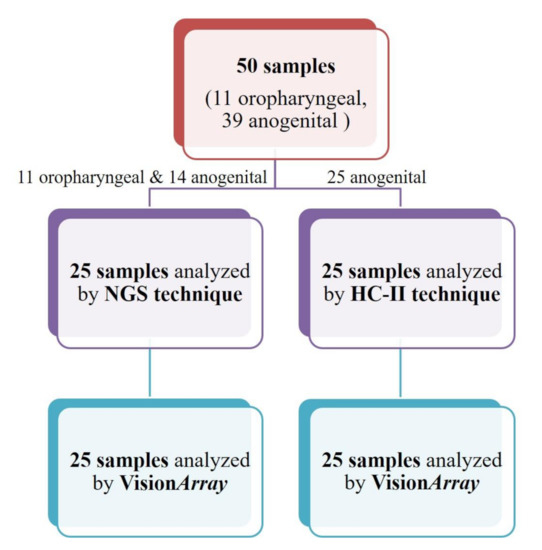
Figure 1.
Flow chart of the analyzed specimens. NGS: Next generation sequencing; HC II: Hybrid Capture II.
Briefly, at least 10 ng of DNA per each PCR reaction were amplified by targeting the L1 gene using fusion primers that were designed starting from a previously described method and modified with multiple identifier (MID) nucleotides: MY09/MY11 (amplicon size: 450 bp) [16] and GP5+/6+ (amplicon size: 150 bp) [34]. Using this system, it is possible to identify the following HPV types: (i) low risk (LR-HPV): 6, 11, 42, 54, and 61; (ii) probably high risk (PHR-HPV): 26, 53, 66, 68, 69, 70, 73, and 82; (iii) high risk (HR-HPV): 16, 18, 31, 33, 35, 45, 51, 52, 56, 58, and 59. The following amplicons were used to evaluate DNA integrity: BRAF Ex15 (199 bp), SOD1 Ex1 (160 bp), and TARDBP Ex5 (486 bp). PCR reactions were evaluated using 3% agarose gel. Negative samples were analyzed twice. Positive samples were sequenced to characterize HPV type using a 454 GS-Junior machine, according to established protocols. Obtained sequences were then blasted using the PaVe database (https://pave.niaid.nih.gov/ (accessed on 1 January 2020)) to identify the HPV type (Supplementary Figure S1).
2.4. Hybrid Capture II
Hybrid Capture II (HC II) analysis was performed using the automated HC II test system (Qiagen, Gaithersburg, MD, USA), according to the manufacturer’s instructions. The Qiagen test is able to identify 13 different HR-HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68) (www.qiagen.com (accessed on 1 June 2020)). Twenty-five fresh specimens (S26–S50, Table 1), all from the anogenital region (cervix), were tested by HC II (Table 1, Figure 1).
Specimens were considered positive for HPV infection when the relative lighting unit/cutoff (RLU/CO) ratio was ≥1.0 (Supplementary Table S1), according to the manufacturer’s instructions.
2.5. VisionArray® HPV Chips
All the 50 specimens analyzed by NGS or HC II were also analyzed using the VisionArray® HPV Chips (ZytoVision GmbH, Bremerhaven, Germany) assay (Table 1, Figure 1). Briefly, at least 15 ng of DNA were amplified by PCR, using the CE-IVD VisionArray® HPV Primer Kit (ZytoVision GmbH, Bremerhaven, Germany) and AmpliTaq Gold DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). VisionArray® HPV Primers target to the L1 region of the Human HPV genomes (amplicon length: 139–148 bp) and allow for the identification of the following HPV types: (i) LR-HPV (6, 11, 40, 42, 43, 44, 54, 55, 57, 61, 62, 72, 81CP8304, 83MM7, 84MM8, 90, and 91); (ii) PHR-HPV (26, 34, 53, 66, 67, 68a, 68b, 69, 70, 73, 82IS39, and 82MM4); (iii) HR-HPV (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59). Primers against the human HLA-DQA1 gene (amplicon length: 222 bp) were also included as a DNA integrity control. Obtained amplicons were detected using hybridization between the amplified sequences and the complementary DNA probe captures on a glass chip. The specific bound biotinylated sequences were secondarily labeled with a streptavidin–peroxidase conjugate afterward and visualized by tetramethylbenzidine (TMB) staining. HPV types were analyzed using the VisionArray® Analyzer Software (ZytoVision GmbH, Bremerhaven, Germany) (Supplementary Figure S2).
2.6. Statistical Analysis
The intra-techniques agreement was calculated using the weighted Cohen kappa graded as complete agreement (κ > 0.81), substantial agreement (κ = 0.61–0.80), moderate agreement (κ = 0.41–0.60), fair agreement (κ = 0.21–0.40), or slight agreement (κ = 0.01–0.20). Statistical performances were calculated using the VassarStats tool (http://vassarstats.net/ (accessed on 1 December 2020)).
3. Results
3.1. Next Generation Sequencing and VisionArray® HPV Chips Analysis Comparison
Thirteen of the 25 samples (52.0%) analyzed using NGS were positive for the presence of the HPV genome, 9 (36.0%) were negative, and 3 (12.0%) were not evaluable due to low quality DNA (Table 2).

Table 2.
Next generation sequencing and VisionArray® HPV Chips analysis comparison.
Considering only the positive cases, 5 were LR-HPV (38.5%), 7 were HR-HPV (53.8%), and 1 was PHR-HPV (7.7%). In 3 of the 13 (23.1%) positive cases, a co-infection by two different HPV types was observed.
The same 25 samples were also analyzed using the VisionArray® HPV kit (Table 2). Sixteen of the 25 samples (64.0%) analyzed were HPV-positive, 9 (36.0%) were negative.
Considering only the positive cases, 6 were LR-HPV (37.5%), 9 were HR-HPV (56.3%), and 1 was PHR-HPV (6.2%). In 5 of the 16 (31.2%) positive cases, HPV co-infection was observed.
3.1.1. Discrepant Results
Comparing NGS to VisionArray®, we observed six cases with discrepant results (Table 2). Three cases (S4, S5, S7, Table 2) were not evaluable in NGS analysis but using the VisionArray® assay they showed one as negative, one as positive for co-infection by HPV types 16 and 42, and one positive for HPV type 16 (Table 2).
One sample (S21, Table 2) was negative if analyzed with NGS, while it was positive for HPV type 40 using VisionArray®.
One sample (S6, Table 2) was detected as co-infected by HPV types 6 and 66 using NGS; the co-infection was also detected by VisionArray® analysis, but other than HPV6 and HPV66, 4 different HPV types were also identified (61, 73, 82, 90, Table 2).
Sample S22 (Table 2), diagnosed as positive for HPV type 35 using NGS, was diagnosed as co-infected with HPV types 34 and 35 using VisionArray®.
3.1.2. Statistical Measures of Performance
A substantial agreement (κ = 0.702, 95% C.I. 0.44–0.97) between NGS and VisionArray® was reached, with an accuracy of 0.84, sensitivity equal to 81.0%, and specificity of 100% (Table 3).

Table 3.
Comparison between next generation sequencing and the VisionArray® HPV Chips techniques.
When only HR-HPV cases were considered as positive, sensitivity, specificity, accuracy, and Cohen’s k were equal to 0.779 (Table 3). If only HR and PHR cases were evaluated as positive, Cohen’s κ was 0.775, accuracy was 0.88, sensitivity 73%, and specificity was 100%. If HR, PHR, and LR cases were considered separately, an optimal concordance (κ = 0.818) was reached (Table 3).
If only evaluable data were considered, a complete agreement (κ = 0.90, 95% C.I. 0.72–1.0) between NGS and VisionArray® was reached, with an accuracy of 0.955, sensitivity equal to 92.9%, and the specificity was 100%. Moreover, when only HR-HPV cases were considered as positive, sensitivity, specificity, accuracy, and Cohen’s k between NGS and VisionArray® were equal to 1.0 (Table 3).
3.2. Hybrid Capture II High-Risk HPV DNA Analysis and VisionArray® HPV Chips Analysis Comparison
Using the HC II test, all 25 samples were evaluable. All of them were from anogenital lesions (Table 1). The HPV genome was detected in 12 of 25 (48.0%) samples, while 13 specimens (52.0%) were negative for the HPV genome (Table 4 and Supplementary Table S1).

Table 4.
Hybrid Capture II and VisionArray® HPV Chips analysis comparison.
The same 25 samples were also analyzed using the VisionArray® HPV kit (Table 3). The HPV genome was detected in 17 of 25 (68.0%) samples, while 8 specimens (32.0%) were negative for the HPV genome Considering only the positive cases, 4 of the 17 were LR-HPV (23.5%), 12 were HR-HPV (70.6%), and 1 was PHR-HPV (5.9%). In 8 of the 16 (50%) positive cases, HPV co-infection was observed (Table 4).
3.2.1. Discrepant Results
By comparing HC II HPV analysis to VisionArray® we observed six cases with different results (S26, S27, S28, S30, S31, and S49, Table 4). Five of them were negative according to the HC II assay, while VisionArray® analysis detected the following HPV types: HPV-42 (S26); HPV-55 (S27); HPV-42 (S28); HPV-16 and HPV-42 (S30); and HPV-6, HPV-11, and HPV-53 (S31) (Table 4). Sample S49 was positive according to Hybrid Capture II but showed infection by low-risk HPV typed according to the VisionArray® analysis (Table 4).
3.2.2. Statistical Measures of Performance
A substantial agreement was reached between VisionArray® and HC (κ = 0.606, 95% C.I. 0.32–0.89). The accuracy was 0.80, the sensitivity 70.6%, and the specificity was 100% (Table 5). When only high-risk HPV cases were considered as positive, sensitivity, specificity, and accuracy were 91.7%, 92.3%, 0.92, respectively. The concordance reached a complete agreement (κ = 0.84). If HR and PHR cases were considered as positive, a substantial agreement was obtained (κ = 0.760), overall accuracy was 0.88, the sensitivity 85%, and the specificity was 92% (Table 5).

Table 5.
Comparison between the Hybrid Capture II analysis and VisionArray® HPV Chips techniques.
4. Discussion
HPV infection is a causative agent involved in the development of about 5% of all human cancers [2,3,4]. The choice of the appropriate test for the diagnosis of HPV infection is very important for the prevention and prognosis of carcinomas in the anogenital and oropharyngeal areas.
In the present study, we analyzed 50 samples from anogenital and oropharyngeal lesions. Of these 50 samples, 25 were tested using both next generation sequencing and the VisionArray® HPV Kit and the other 25 were tested with the Hybrid Capture® 2 High-Risk HPV DNA Test and the VisionArray® HPV Kit.
Three specimens (S4, S5, and S7) were not evaluable with the NGS technique while with VisionArray® one sample showed negative and two samples positive. This discrepancy could be due to the different length of amplicons sequenced by NGS and those analyzed using the VisionArray® HPV Kit. All these samples were FFPE specimens with consequent DNA degradation due to the formalin fixation. The evidence that amplicons of the VisionArray® HPV Kit are shorter than those analyzed by NGS may be the reason for the discrepancy because these three samples were evaluable by the VisionArray® HPV Kit but not by NGS.
One sample (S21) was negative when analyzed with the NGS platform and HPV- positive (HPV-40) using VisionArray® HPV Kit. The sample S22 was positive for HPV-35 (HR-HPV) using NGS analysis, but it showed as co-infected by both HPV-34 (PHR-HPV) and HPV-35 when analyzed with the VisionArray® assay. However, it should be considered that the primers used for NGS analysis were not able to identify HPV-34. In this study, to detect HPV using a PCR-based NGS assay, degenerate primers were used. These primers provide an alternative to the use of mixtures of type-specific primers. However, HPV-6, HPV-16, HPV-18, HPV-31, HPV-33, and HPV-35 provide more preferred templates for the consensus PCR primers than those of other genotypes [35]. In sample 22, HPV-34 was found together with HPV-35, leading to no detection of HPV-34 by NGS.
One sample (S6) was detected as co-infected by HPV types 6 and 66 using 454 GS-Junior; the co-infection was also detected by VisionArray® analysis, but four different HPV types were identified other than 6 and 66: HPV-61, -73, -82, -90). This discrepancy might be because HPV-61 (LR), -73 (PHR), and -82 (PHR) sequences were “trimmed” by the NGS software due to low-quality values, while HPV-90 (LR) is not detectable by the primers used for NGS analysis. As reported above, HPV-6, HPV-16, HPV-18, HPV-31, HPV-33, and HPV-35 provide more preferred templates for the consensus PCR primers [35]. In sample 6, HPV-90 was found together with several other HPV types including HPV-6, and so it is possible to hypothesize that HPV-90 is not detected by NGS.
Six cases showed different results when HC II and VisionArray assays were compared. Five of them (S26, S27, S28, S31, and S30) were negative according to the HC II assay, but VisionArray® identified several HPV types. Samples S26 (HPV-42, LR), S27 (HPV-55, LR), S28 (HPV-42, LR), and S31 (HPV-53, PHR) were positive for LR- or PHR-HPVs not detectable by the HC II test. The S30 sample (HPV-16–HR and HPV-42–LR), was diagnosed as negative with the HC II method. This HC negative result might be due to a very low level of infection, and for this reason was evaluated as “negative” using HC II. The limit of detection of HC II was 5000 HPV copies per test (according to the manufacturer’s instruction), while it was from 50 to 5000 genome equivalents (GEM) per test (500,000 GEM for HPV68a) using VisionArray® (according to the manufacturer’s instruction), and about 100 HPV copies per test using PCR based NGS [31,32]. The sample S49 was positive according to HC II but showed a low-risk HPV type (HPV40+42+44+54) according to VisionArray® analysis. The analysis was repeated twice and gave the same results. Not enough material was available for testing this sample with NGS. It could be possible that the presence of multiple HPV types in the analyzed sample leads to the preferential amplification of the HPV types detected by VisionArray® (probably because they are more abundant), “losing” the HR-HPV type DNA detected by HC II (not designed for LR-HPV identification).
The VisionArray® methodology presented a substantial concordance if compared with the NGS technique because both systems identified almost the same HPV types. The statistical measurement of performance was influenced by four cases: three that were not evaluable by NGS but positive (2 cases) or negative (1 case) by VisionArray®, and one that was negative by NGS and positive (HPV40–LR-HPV) by VisionArray®. All the three not-evaluable cases by NGS were from the FFPE specimens, leading us to hypothesize a better performance of VisionArray® starting from formalin-fixed specimens. Moreover, it should be considered that NGS performed worse if lesions presented multiple HPV infections. This could be because, during the sequences analysis, the obtained sequences must be blasted using an online database to identify the HPV type; this step could lead to losing the HPV type that is less represented in the specimen analyzed. In this study, a target amplicon-based panel was used. The target amplicon panels need a PCR before the NGS protocol and are typically more sensitive than NGS without a previous PCR. However, a large not-amplicon-based panel results in a better sensitivity because it could find mismatches in the sequence of the primers used for PCR amplification [22,25,33]. In this study, we started from at least 10 ng of purified DNA for NGS analysis and from 15 ng for the VisionArray® (as indicated by the manufacturer’s instructions). Provably, the use of a single couple of degenerated primers (NGS) instead of a multiplex of primers (VisionArray®) allows for a slight reduction of the minimum input of needed DNA. It should be considered that the costs of NGS are highly influenced by the number of samples that are analyzed in the same NGS run. If only a few samples are analyzed, the cost of an NGS run is much more expensive than other tests that could be used for HPV typing. In this case, the use of targeted custom panels would allow for the running of multiple panels in the same NGS run, optimizing the cost per sample. Finally, the analysis of the results should be considered: NGS analysis usually requires a bioinformatic pipeline analysis of the output sequences, which is more challenging than working with the data obtained by other kits (e.g., those obtained by HC II or VisionArray®—See Supplementary Material).
For oropharyngeal lesions, if on the one hand it is true that according to the guidelines p16 analysis is a good surrogate for identifying HPV lesions in oropharyngeal carcinomas, it has also been demonstrated that an alternative method parallel to p16 analysis is recommended in these types of lesions [36]. According to our data, NGS or VisionArray® are techniques that could be performed together with p16 immunostaining in oropharyngeal carcinomas. Some studies have found a good correlation between p16 IHC (immunohistochemistry) and NGS HPV analysis [24,37,38], however, further data are needed to deeply analyze the clinical utility of two-tiered (NGS and p16 IHC) HPV testing processes in oropharyngeal carcinomas.
A substantial agreement was obtained between the VisionArray® assay and the HC II assay: It should be considered that the HC II assay is designed to diagnose only HR-HPV types. When only HR genotypes were investigated, HC II assay performed properly. According to our data, NGS and VisionArray® assays resulted in a highly sensitive and specific method to diagnose the presence of HPV DNA both in fresh and FFPE specimens, while other assays, such as HC-II, are not applicable with formalin-fixed material. It is also important to use a method that allows for the detection of HPV co-infection in the analyzed specimens, as stated by the HPV LabNet International Proficiency Study [39]; in fact, the underestimation of the prevalence of multiple infections introduces a systematic bias in epidemiological studies [39]. It should be considered that in a molecular pathology laboratory, the most analyzed specimens are from FFPE material from the anatomic pathology section. For example, HC II assay is enough to detect HR-HPV in anogenital lesions, as also demonstrated by our data, but this method is not optimized for the analysis of FFPE material. For this reason, protocols that can obtain robust results from both fresh-frozen and FFPE samples should be available in a molecular pathology laboratory. The choice of the more appropriate technique for HPV detection should be guided not only by the analytical performance, but also by the clinical aims, starting material, costs, and turnaround time.
Supplementary Materials
The following are available online at https://www.mdpi.com/2673-5261/2/1/4/s1, Figure S1: Example of HPV positive sample (S16 Table 1) after NGS sequencing and alignment on PaVe database, Figure S2: Example of HPV positive (A, S16 Table 1) and negative (B, S3 Table 1) specimens obtained by VisionArray®, Table S1: Results of samples analyzed by HC II and VisionArray.
Author Contributions
Conceptualization, G.A., M.V., and D.d.B.; methodology, G.A., M.V., V.S., T.M., C.C.O., A.D.L., P.P., and P.C.; formal analysis, G.A., M.V., and P.P.; investigation, data curation, G.A., M.V., and D.d.B.; writing—original draft preparation, G.A. and M.V.; writing—review and editing, G.C., A.P., G.T., and D.d.B.; supervision, A.P., G.T., and D.d.B.; project administration, A.P. and G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All the experimental procedures were carried out in accordance with the general authorization to process personal data for scientific research purposes from “The Italian Data Protection Authority” (http://www.garanteprivacy.it/web/guest/home/docweb/-/docwebdisplay/export/2485392 (accessed on 7 January 2021)). All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Helsinki Declaration (https://www.wma.net/fr/news-post/en-matierede-transfert-des-taches-la-securite-des-patients-et-la-qualitedes-soins-devraient-etre-primordiales/ (accessed on 7 January 2021)). The study did not affect the clinical management of the involved patients’ samples.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schiller, J.T.; Day, P.M.; Kines, R.C. Current understanding of the mechanism of HPV infection. Gynecol. Oncol. 2010, 118, S12–S17. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615. [Google Scholar] [CrossRef]
- Forman, D.; de Martel, C.; Lacey, C.J.; Soerjomataram, I.; Lortet-Tieulent, J.; Bruni, L.; Vignat, J.; Ferlay, J.; Bray, F.; Plummer, M.; et al. Global burden of human papillomavirus and related diseases. Vaccine 2012, 30 (Suppl. 5), F12–F23. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H. The search for infectious causes of human cancers: Where and why (Nobel lecture). Angew. Chem. Int. Ed. Engl. 2009, 48, 5798–5808. [Google Scholar] [CrossRef]
- Munoz, N.; Bosch, F.X.; de Sanjose, S.; Herrero, R.; Castellsague, X.; Shah, K.V.; Snijders, P.J.; Meijer, C.J. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- de Sanjose, S.; Diaz, M.; Castellsague, X.; Clifford, G.; Bruni, L.; Munoz, N.; Bosch, F.X. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: A meta-analysis. Lancet Infect. Dis. 2007, 7, 453–459. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens--Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- Jacobs, M.V.; de Roda Husman, A.M.; van den Brule, A.J.; Snijders, P.J.; Meijer, C.J.; Walboomers, J.M. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J. Clin. Microbiol. 1995, 33, 901–905. [Google Scholar] [CrossRef]
- van den Brule, A.J.; Pol, R.; Fransen-Daalmeijer, N.; Schouls, L.M.; Meijer, C.J.; Snijders, P.J. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 2002, 40, 779–787. [Google Scholar] [CrossRef]
- Davies, P.; Kornegay, J.; Iftner, T. Current methods of testing for human papillomavirus. Best Pract. Res. Clin. Obstet. Gynaecol. 2001, 15, 677–700. [Google Scholar] [CrossRef]
- Gravitt, P.E.; Peyton, C.L.; Apple, R.J.; Wheeler, C.M. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 1998, 36, 3020–3027. [Google Scholar] [CrossRef]
- Human papillomavirus vaccines: WHO position paper, May 2017-Recommendations. Vaccine 2017, 35, 5753–5755. [CrossRef] [PubMed]
- Franceschi, S.; Herrero, R.; Clifford, G.M.; Snijders, P.J.; Arslan, A.; Anh, P.T.; Bosch, F.X.; Ferreccio, C.; Hieu, N.T.; Lazcano-Ponce, E.; et al. Variations in the age-specific curves of human papillomavirus prevalence in women worldwide. Int. J. Cancer 2006, 119, 2677–2684. [Google Scholar] [CrossRef] [PubMed]
- Rischin, D.; Young, R.J.; Fisher, R.; Fox, S.B.; Le, Q.T.; Peters, L.J.; Solomon, B.; Choi, J.; O’Sullivan, B.; Kenny, L.M.; et al. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 4142–4148. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Manos, M.M.; Ting, Y.; Wright, D.K.; Lewis, J.; Broker, T.R.; Wolinsky, S.M. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells 1989, 7, 209–214. [Google Scholar]
- Coutlee, F.; Gravitt, P.; Kornegay, J.; Hankins, C.; Richardson, H.; Lapointe, N.; Voyer, H.; Franco, E. Use of PGMY primers in L1 consensus PCR improves detection of human papillomavirus DNA in genital samples. J. Clin. Microbiol. 2002, 40, 902–907. [Google Scholar] [CrossRef]
- Snijders, P.J.; van den Brule, A.J.; Schrijnemakers, H.F.; Snow, G.; Meijer, C.J.; Walboomers, J.M. The use of general primers in the polymerase chain reaction permits the detection of a broad spectrum of human papillomavirus genotypes. J. Gen. Virol. 1990, 71 Pt 1, 173–181. [Google Scholar] [CrossRef]
- Fontaine, V.; Mascaux, C.; Weyn, C.; Bernis, A.; Celio, N.; Lefevre, P.; Kaufman, L.; Garbar, C. Evaluation of combined general primer-mediated PCR sequencing and type-specific PCR strategies for determination of human papillomavirus genotypes in cervical cell specimens. J. Clin. Microbiol. 2007, 45, 928–934. [Google Scholar] [CrossRef]
- Cullen, M.; Boland, J.F.; Schiffman, M.; Zhang, X.; Wentzensen, N.; Yang, Q.; Chen, Z.; Yu, K.; Mitchell, J.; Roberson, D.; et al. Deep sequencing of HPV16 genomes: A new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res. 2015, 1, 3–11. [Google Scholar] [CrossRef]
- Lee, J.Y.; Cutts, R.J.; White, I.; Augustin, Y.; Garcia-Murillas, I.; Fenwick, K.; Matthews, N.; Turner, N.C.; Harrington, K.; Gilbert, D.C.; et al. Next Generation Sequencing Assay for Detection of Circulating HPV DNA (cHPV-DNA) in Patients Undergoing Radical (Chemo)Radiotherapy in Anal Squamous Cell Carcinoma (ASCC). Front. Oncol. 2020, 10, 505. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Unger, E.R.; Rajeevan, M.S. Universal human papillomavirus typing by whole genome sequencing following target enrichment: Evaluation of assay reproducibility and limit of detection. BMC Genom. 2019, 20, 231. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Unger, E.R.; Batra, D.; Sheth, M.; Steinau, M.; Jasinski, J.; Jones, J.; Rajeevan, M.S. Universal Human Papillomavirus Typing Assay: Whole-Genome Sequencing following Target Enrichment. J. Clin. Microbiol. 2017, 55, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, N.D.; Parker, J.S.; Eberhard, D.A.; Patel, N.M.; Weck, K.E.; Sharpless, N.E.; Hu, Z.; Hayes, D.N.; Gulley, M.L. Identification of Human Papillomavirus Infection in Cancer Tissue by Targeted Next-generation Sequencing. Appl. Immunohistochem. Mol. Morphol. 2016, 24, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Kocjan, B.J.; Bzhalava, D.; Forslund, O.; Dillner, J.; Poljak, M. Molecular methods for identification and characterization of novel papillomaviruses. Clin. Microbiol. Infect. 2015, 21, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Chandrani, P.; Kulkarni, V.; Iyer, P.; Upadhyay, P.; Chaubal, R.; Das, P.; Mulherkar, R.; Singh, R.; Dutt, A. NGS-based approach to determine the presence of HPV and their sites of integration in human cancer genome. Br. J. Cancer 2015, 112, 1958–1965. [Google Scholar] [CrossRef] [PubMed]
- Nowak, R.G.; Ambulos, N.P.; Schumaker, L.M.; Mathias, T.J.; White, R.A.; Troyer, J.; Wells, D.; Charurat, M.E.; Bentzen, S.M.; Cullen, K.J. Genotyping of high-risk anal human papillomavirus (HPV): Ion torrent-next generation sequencing vs. linear array. Virol. J. 2017, 14, 112. [Google Scholar] [CrossRef]
- Nilyanimit, P.; Chansaenroj, J.; Poomipak, W.; Praianantathavorn, K.; Payungporn, S.; Poovorawan, Y. Comparison of Four Human Papillomavirus Genotyping Methods: Next-generation Sequencing, INNO-LiPA, Electrochemical DNA Chip, and Nested-PCR. Ann. Lab. Med. 2018, 38, 139–146. [Google Scholar] [CrossRef]
- Flores-Miramontes, M.G.; Torres-Reyes, L.A.; Alvarado-Ruiz, L.; Romero-Martinez, S.A.; Ramirez-Rodriguez, V.; Balderas-Pena, L.M.; Vallejo-Ruiz, V.; Pina-Sanchez, P.; Cortes-Gutierrez, E.I.; Jave-Suarez, L.F.; et al. Human papillomavirus genotyping by Linear Array and Next-Generation Sequencing in cervical samples from Western Mexico. Virol. J. 2015, 12, 161. [Google Scholar] [CrossRef]
- Dias, T.C.; Longatto-Filho, A.; Campanella, N.C. Human papillomavirus genotyping as a tool for cervical cancer prevention: From commercially available human papillomavirus DNA test to next-generation sequencing. Future Sci. OA 2020, 6, FSO603. [Google Scholar] [CrossRef]
- Barzon, L.; Militello, V.; Lavezzo, E.; Franchin, E.; Peta, E.; Squarzon, L.; Trevisan, M.; Pagni, S.; Dal Bello, F.; Toppo, S.; et al. Human papillomavirus genotyping by 454 next generation sequencing technology. J. Clin. Virol. 2011, 52, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, L.S.; Smelov, V.; Bzhalava, D.; Eklund, C.; Hultin, E.; Dillner, J. Next generation sequencing for human papillomavirus genotyping. J. Clin. Virol. 2013, 58, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Arroyo Muhr, L.S.; Hultin, E.; Bzhalava, D.; Eklund, C.; Lagheden, C.; Ekstrom, J.; Johansson, H.; Forslund, O.; Dillner, J. Human papillomavirus type 197 is commonly present in skin tumors. Int. J. Cancer 2015, 136, 2546–2555. [Google Scholar] [CrossRef]
- de Roda Husman, A.M.; Walboomers, J.M.; van den Brule, A.J.; Meijer, C.J.; Snijders, P.J. The use of general primers GP5 and GP6 elongated at their 3’ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995, 76 Pt 4, 1057–1062. [Google Scholar] [CrossRef]
- Lee, S.H.; Vigliotti, V.S.; Vigliotti, J.S.; Pappu, S. Validation of human papillomavirus genotyping by signature DNA sequence analysis. BMC Clin. Pathol. 2009, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.G.; Anderson, L.A.; Schache, A.G.; Moran, M.; Graham, L.; Currie, K.; Rooney, K.; Robinson, M.; Upile, N.S.; Brooker, R.; et al. Recommendations for determining HPV status in patients with oropharyngeal cancers under TNM8 guidelines: A two-tier approach. Br. J. Cancer 2019, 120, 827–833. [Google Scholar] [CrossRef]
- Ambulos, N.P., Jr.; Schumaker, L.M.; Mathias, T.J.; White, R.; Troyer, J.; Wells, D.; Cullen, K.J. Next-Generation Sequencing-Based HPV Genotyping Assay Validated in Formalin-Fixed, Paraffin-Embedded Oropharyngeal and Cervical Cancer Specimens. J. Biomol. Tech. 2016, 27, 46–52. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Conway, C.; Chalkley, R.; High, A.; Maclennan, K.; Berri, S.; Chengot, P.; Alsop, M.; Egan, P.; Morgan, J.; Taylor, G.R.; et al. Next-generation sequencing for simultaneous determination of human papillomavirus load, subtype, and associated genomic copy number changes in tumors. J. Mol. Diagn. 2012, 14, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Eklund, C.; Forslund, O.; Wallin, K.L.; Dillner, J. Global improvement in genotyping of human papillomavirus DNA: The 2011 HPV LabNet International Proficiency Study. J. Clin. Microbiol. 2014, 52, 449–459. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).