1. Introduction
In the practice of forest resource field sample surveys, tree species is an essential survey factor. Tree species information is also an important parameter for ecosystem modeling and forest resource management [
1]. With the continuous development of light detection and ranging (LiDAR) technology, the 3D structural parameters of forest trees can be obtained quickly and accurately by using ground-based LiDAR systems for sample plot scanning. The focus and difficulty of current research is how to accurately identify tree species information from laser point clouds of individual trees.
Traditional machine learning methods necessitate the manual extraction of copious amounts of 3D structural information for modeling, and recognition accuracy is low [
2,
3,
4,
5,
6]. A breakthrough in computer vision has been made in classifying 3D object shapes using point cloud deep learning techniques, opening up a new practical direction for tree species classification.
Some previous studies converted LiDAR data into images with various classification features [
7,
8,
9] and then used image deep learning methods to study the classification of tree species. With the development of point-based deep learning methods such as PointNet [
10] and PointNet++ [
11], scholars have started to use point-by-point deep learning models for tree species classification research. Seidel et al. [
12] used PointNet to classify seven tree species, but the classification accuracy was very low and then shifted the focus of the study to a picture convolutional neural network (CNN) approach, which finally achieved a high classification accuracy. In other studies, using point cloud deep learning models [
13,
14,
15,
16,
17,
18,
19], good accuracy of tree classification was achieved.
To explore the potential of point cloud deep learning for individual tree species point cloud classification, we used three pointwise MLP-based point cloud deep learning methods (PointNet, PointNet++, PointMLP) to classify tree species from individual tree point clouds of four and seven tree species. We used the farthest point sampling method to reduce the number of points in each individual tree point cloud to 1024 and 2048. We achieved extremely exciting experimental results. In the classification experiments with a balanced sample size, PointMLP and PointNet++ achieved high accuracy in tree classification on the test set.
2. Materials and Methods
2.1. Individual Tree Point Cloud Data
In this study, we used a publicly available dataset for our experiments [
20]. The name of the dataset is “Single tree point clouds from terrestrial laser scanning”. It contains individual tree point cloud data for seven tree species (
Table 1). Detailed information about the data acquisition and sensors can be found in the article [
12].
2.2. Data Preprocessing
We referred to the [
12] process for data preprocessing with some modifications. We performed the following four preprocessing operations on the point cloud data.
Manual selection: The individual tree data were screened artificially to control the quality of the individual tree samples.
Deleted: Deleted the point of 30% density at the bottom of the tree.
Downsampling: The farthest point sampling method was used to sample the points of the individual tree as 1024 and 2048.
Data organization: The data organization of the ModelNet40 [
21] dataset was used to create the sample database for this study experiment.
2.3. Point Cloud Deep Learning Models
PointNet [
10] is the pioneer of point-based deep learning. PointNet uses a shared MLP to directly process unordered point sets as input. PointNet contains three key modules: a maximum pooling layer as a symmetric function to aggregate information from all points, a combined local and global information structure, and two joint alignment networks for aligning input points and point features.
PointNet is unable to capture the local structure generated by metric space points, thus limiting its ability to recognize fine-grained patterns and generalize to complex scenes. The proposed PointNet++ [
11] method bridges this gap. PointNet++ is a hierarchical neural network that addresses two core problems: how to generate partitions of point sets and how to abstract point sets or local features by local feature learners. The position and scale of the prime points are used to describe each partition of the click, and the FPS algorithm is used to select the prime points. Features extracted within each partition using PointNet are used as descriptions of local features. The model structure contains two ensemble abstraction layers (SAs). Each SA layer consists of three parts: a sampling layer, a grouping layer, and a PointNet layer.
The PointMLP [
22] model follows the design philosophy of PointNet, a simpler but deeper network architecture. PointMLP learns the point cloud representation by a simple feed-forward residual MLP network that hierarchically aggregates the local features extracted by the MLP. PointMLP introduces a lightweight local geometric affine module that adaptively transforms point features in local regions. PointMLP is similar to PointNet and PointNet++, but it is more general and exhibits better performance.
The hyperparameters of PointNet, PointNet++, and PointMLP model training are summarized in
Table 2. PyTorch (1.10.0 + CUDA 11.3) is the framework used by three deep learning methods. The graphics card used in this study was an NVIDIA GeForce RTX 3070 (8 GB).
2.4. Model Accuracy Evaluation Metrics
In this experiment, balanced accuracy (BAcc) and
kappa coefficients were used to compare and analyze the results of tree species classification in terms of accuracy evaluation and models.
The kappa coefficient is calculated from the confusion matrix of the classification results, and its value ranges from −1 to 1. Usually, the kappa coefficient is greater than zero, and its absolute value is larger, indicating better classification results.
3. Results
The tree classification results for the six sets of experiments using the three deep learning models are summarized in
Table 3. As seen from the table, the accuracy of the experiments using the four tree species with balanced sample data for classification is relatively high. The experiments with 2048 sampling points achieved a higher accuracy of tree species classification. A comparison of the classification accuracies of the test dataset shows that the results of both models, PointNet++ and PointMLP, are increasingly similar.
By mapping the training accuracy of each epoch during model training (
Figure 1), we portrayed the training process of the three deep learning models. As seen from the figure, the classification accuracy of the PointNet model is extremely low, and the classification accuracy of the test dataset does not increase with the increasing training accuracy of the model on the test dataset. During the operation of the PointNet++ model, the classification accuracy of the test set was higher than that of the training set and has been growing, finally obtaining a high classification accuracy. The test accuracy of the PointMLP model increases with the training accuracy and finally achieves a classification accuracy similar to that of the PointNet++ model.
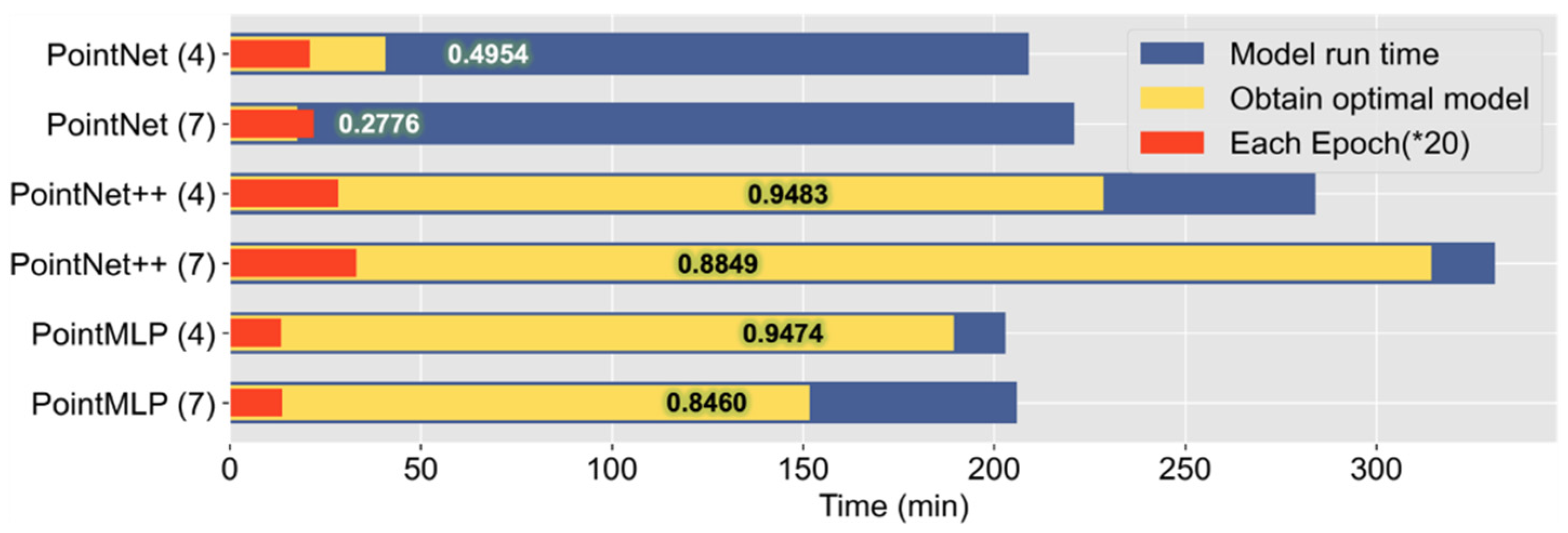
We summarized and plotted the time trained for the six sets of experiments with 2048 sampling points (
Figure 2). It can be seen from the figure that the PointNet++ model is more time-consuming, and it takes a longer time to train to obtain the optimal model parameters, while the PointMLP model can achieve a classification accuracy similar to that of the PointNet++ model in a brief period. The PointNet model saturates the model performance shortly after starting training, and the final classification accuracy is extremely low.
4. Discussion
In this study, three pointwise MLP-based point cloud deep learning methods were used to investigate the classification of individual tree point clouds. According to the findings, two deep learning models, PointNet++ and PointMLP, can better identify the tree species information in individual tree point clouds.
The tree species classification obtained by the PointNet model in our study was exceptionally low, similar to the results obtained by Seidel et al. [
12]. This is because PointNet cannot capture local features of 3D objects, which limits its ability to classify and recognize similar objects. However, both the PointNet++ and PointMLP models introduce a local feature extraction module, which can extract the fine-grained local features of 3D objects well and thus achieve good classification accuracy. A related study [
19] also demonstrated that PointNet++ can achieve good classification accuracy for tree species classification problems.
In this study, the experiments to classify the four tree species achieved a high classification accuracy. This is because the four tree species used have a larger number of samples and are more balanced in number. The other three tree species have a smaller number of samples, which limits the further learning of classification features by deep learning. From this, we suggest that when using deep learning methods for object classification in the future, researchers try to choose a substantial number of samples and keep the distribution of the number of samples consistent.
The classification accuracy is higher when the number of sampling points of an individual tree is 2048, which is consistent with the findings of [
19]. This indicates that 1024 points are not a good representation of the accurate 3D structural information of individual trees.
Our experiments show that the two models, PointNet++ and PointMLP, achieve comparable tree classification accuracy. However, the PointMLP model can be trained in less time to obtain the optimal deep learning model parameters. This is due to PointMLP’s simpler and deeper network architecture, which is achieved by learning the point cloud representation using a simple feed-forward residual MLP network. PointMLP has exceptional model performance.
5. Conclusions
Point cloud deep learning models of the MLP type with local feature extraction are proven to be accurate for tree species classification of individual tree point clouds. PointMLP, the current state-of-the-art (SOTA) MLP-based point cloud deep learning method, has promising applications in tree species classification.
Author Contributions
Conceptualization, B.L.; methodology, B.L.; software, M.R.; validation, B.L. and M.R.; formal analysis, B.L.; investigation, B.L.; resources, B.L.; data curation, B.L. and M.R.; writing—original draft preparation, B.L.; writing—review and editing, H.H. and X.T.; visualization, B.L.; supervision, H.H.; project administration, X.T. and H.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant number: 42130111 and 41930111), the National Key Research and Development Program of China (Grant number: 2021YFE0117700), and the Fundamental Research Funds of CAF (Grant number: CAFYBB2021SY006).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Terryn, L.; Calders, K.; Disney, M.; Origo, N.; Malhi, Y.; Newnham, G.; Raumonen, P.; Å kerblom, M.; Verbeeck, H. Tree species classification using structural features derived from terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 170–181. [Google Scholar] [CrossRef]
- Åkerblom, M.; Raumonen, P.; Mäkipää, R.; Kaasalainen, M. Automatic tree species recognition with quantitative structure models. Remote Sens. Environ. 2017, 191, 1–12. [Google Scholar] [CrossRef]
- Ba, A.; Laslier, M.; Dufour, S.; Hubert-Moy, L. Riparian trees genera identification based on leaf-on/leaf-off airborne laser scanner data and machine learning classifiers in northern France. Int. J. Remote Sens. 2019, 41, 1645–1667. [Google Scholar] [CrossRef]
- Lin, Y.; Herold, M. Tree species classification based on explicit tree structure feature parameters derived from static terrestrial laser scanning data. Agric. For. Meteorol. 2016, 216, 105–114. [Google Scholar] [CrossRef]
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.-A. Identifying the genus or species of individual trees using a three-wavelength airborne lidar system. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, H.; Hu, Q.; Liu, H.; Liu, L.; Bennamoun, M. Deep Learning for 3D Point Clouds: A Survey. IEEE Trans Pattern Anal Mach Intell 2021, 43, 4338–4364. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, B.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171–172, 104–114. [Google Scholar] [CrossRef]
- Zou, X.; Cheng, M.; Wang, C.; Xia, Y.; Li, J. Tree Classification in Complex Forest Point Clouds Based on Deep Learning. IEEE Geosci. Remote Sens. Lett. 2017, 14, 2360–2364. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Kaichun, M.; Guibas, L.J. PointNet: Deep Learning on Point Sets for 3D Classification and Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. PointNet++: Deep Hierarchical Feature Learning on Point Sets in a Metric Space. Adv. Neural Inf. Process. Syst. 2017, 30, 5105–5114. [Google Scholar]
- Seidel, D.; Annighofer, P.; Thielman, A.; Seifert, Q.E.; Thauer, J.H.; Glatthorn, J.; Ehbrecht, M.; Kneib, T.; Ammer, C. Predicting Tree Species From 3D Laser Scanning Point Clouds Using Deep Learning. Front Plant Sci 2021, 12, 635440. [Google Scholar] [CrossRef] [PubMed]
- Briechle, S.; Krzystek, P.; Vosselman, G. Semantic Labeling of Als Point Clouds for Tree Species Mapping Using the Deep Neural Network Pointnet++. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W13, 951–955. [Google Scholar] [CrossRef]
- Briechle, S.; Krzystek, P.; Vosselman, G. Classification of Tree Species and Standing Dead Trees by Fusing Uav-Based Lidar Data and Multispectral Imagery in the 3d Deep Neural Network Pointnet++. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, V-2-2020, 203–210. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C.; Rood, S.B.; Peddle, D.R. See the forest and the trees: Effective machine and deep learning algorithms for wood filtering and tree species classification from terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 1–16. [Google Scholar] [CrossRef]
- Liu, M.; Han, Z.; Chen, Y.; Liu, Z.; Han, Y. Tree species classification of LiDAR data based on 3D deep learning. Measurement 2021, 177, 109301. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Y.; Liu, Z. Classification of Typical Tree Species in Laser Point Cloud Based on Deep Learning. Remote Sens. 2021, 13, 4750. [Google Scholar] [CrossRef]
- Lv, Y.; Zhang, Y.; Dong, S.; Yang, L.; Zhang, Z.; Li, Z.; Hu, S. A Convex Hull-Based Feature Descriptor for Learning Tree Species Classification From ALS Point Clouds. IEEE Geosci. Remote Sens. Lett. 2022, 19, 1–5. [Google Scholar] [CrossRef]
- Liu, B.; Chen, S.; Huang, H.; Tian, X. Tree Species Classification of Backpack Laser Scanning Data Using the PointNet++ Point Cloud Deep Learning Method. Remote Sens. 2022, 14, 3809. [Google Scholar] [CrossRef]
- Seidel, D. Single Tree Point Clouds from Terrestrial Laser Scanning; V2; GRO.data: Göttingen, Germany, 2020. [Google Scholar] [CrossRef]
- Wu, Z.; Song, S.; Khosla, A.; Yu, F.; Zhang, L.; Tang, X.; Xiao, J. 3D ShapeNets: A deep representation for volumetric shapes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1912–1920. [Google Scholar]
- Ma, X.; Qin, C.; You, H.; Ran, H.; Fu, Y. Rethinking Network Design and Local Geometry in Point Cloud: A Simple Residual MLP Framework. In Proceedings of the International Conference on Learning Representations, Virtual, 25–29 April 2022. [Google Scholar]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).