Abstract
For in vivo application of mRNA therapeutics, the development of mRNA nanocarriers that protect mRNA from enzymatic degradation is needed. While current nanocarrier development focuses on fine-tuning the chemical structure of its components, including lipids and polymers, herein, we propose a novel strategy to design stable mRNA nanocarriers by structuring mRNA inside the nanocarriers. Firstly, several mRNA strands were crosslinked with each other using RNA crosslinkers that hybridize to mRNA strands, to prepare mRNA nanoassemblies (NAs). Then, we mixed NAs with poly(ethylene glycol) (PEG)-polycation block copolymers to prepare core–shell-structured polyplex micelles (PMs), composed of PEG shell and mRNA-containing core. Notably, PM-loading NAs (NA/m) exhibited enhanced stability against enzymatic attack and polyion exchange reaction compared to that loading naïve mRNA (naïve/m). According to mechanistic analyses, NA/m possessed a shell with a denser PEG layer and a core with more condensed mRNA compared to naïve/m. As a result, NA/m induced more efficient protein expression after introduction to cultured cells and mouse brain, compared to naïve/m. While newly developed materials need long processes before their clinical approval, our strategy is effective in improving stability and the mRNA introduction efficiency of existing mRNA nanocarriers just by structuring mRNA without the use of additional materials.
1. Introduction
In vitro transcribed messenger RNA (mRNA) garners much attention in next-generation therapeutics, especially in its application to vaccination against cancer and pandemic, protein replacement therapy and genome editing [1,2,3,4]. Owing to the early onset of protein expression from mRNA, acute diseases are also promising targets of mRNA therapeutics [5,6]. Meanwhile, for the wide-spread clinical application of mRNA, there still remains challenges in the delivery technology. Notably, mRNA is more susceptible to enzymatic degradation compared to DNA [7,8], which motivates many researchers to develop mRNA nanocarriers using lipids and polymers [9,10,11]. While such research focusses on modulating the chemical structure of lipids and polymers, additional strategies that work cooperatively with nanocarrier development are required to tackle this challenging issue.
For siRNA therapeutics, chemical modification is performed to increase nuclease stability, which allows in vivo siRNA delivery even without the use of delivery nanocarriers [12]. Chemical modification was also performed in mRNA therapeutics, successfully alleviating the immunogenicity of mRNA [13,14]. However, reported modification formulations allowed only modest improvement in mRNA nuclease stability [15]. Although sugar and phosphate linkage are frequently modified to improve siRNA nuclease stability, most of the targets of mRNA modifications are bases, with very few reports modifying the sugar–phosphate backbone [16], presumably because even subtle modification decreases protein translational activity of mRNA [15,17]. To expand the variety of mRNA modification, we conceived to hybridize mRNA with RNA oligonucleotides possessing stabilizing moieties (Figure 1a,b). Although the hybridization of mRNA with 23 nt or longer RNA oligonucleotides resulted in reduced translational activity and increased immunogenicity of mRNA, 17 nt RNA oligonucleotide did not influence such properties of mRNA [18]. Using this strategy, we introduced cholesterol moieties to mRNA to stabilize polymer-based mRNA nanocarriers [18], and poly(ethylene glycol) (PEG) to mRNA to prevent the aggregation of lipid-based mRNA nanocarriers during their preparation and after their in vivo administration [19]. These reports showed the utility and versatility of our mRNA architectonics approach.
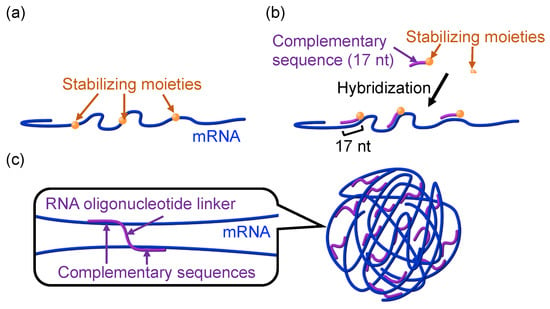
Figure 1.
mRNA architectonics. (a) Chemical modification of mRNA molecules. This method can affect the protein translational activity of mRNA, depending on modification formulations. (b) mRNA engineering using complementary RNA oligonucleotides. By controlling hybridization to be 17 nt, this method allows for the introduction of stabilizing moieties, such as cholesterol and PEG, without reducing mRNA translational activity and increasing mRNA immunogenicity. (c) mRNA nanoassemblies (NA). Several RNA strands were bundled using RNA oligonucleotide linkers possessing sequences complementary to mRNA at their 5′ and 3′ ends. Reproduced from reference [20] with permission.
Along with chemical modification, the modulation of the nucleic acid steric structure is an effective method to improve the nuclease stability of DNA and RNA [21,22,23,24]. Utilizing the mRNA architectonics approach, we crosslinked several mRNA strands using RNA oligonucleotide linkers possessing two sequences complementary to mRNA (Figure 1c). The mRNA nanoassemblies (NAs) thus prepared showed enhanced stability against nucleases compared to naïve mRNA [25]. Interestingly, NAs preserved their translational activity, presumably because NAs may dissociate specifically in cytosol through 5′ cap-dependent translation processes.
Herein, to further improve the functionalities of NAs, we encapsulated NAs into polyplex micelles (PMs), possessing a core–shell structure of a PEG shell and mRNA containing core (see Figure 2). PM is a promising platform of mRNA nanocarriers, effectively preventing mRNA degradation from nucleases and alleviating mRNA immunogenicity by inhibiting mRNA recognition by Toll-like receptors [26,27]. In the present study, PM-loading NAs (NA/m) exhibited enhanced biological and physical stability compared to that loading naïve mRNA (naïve/m), eventually showing increased mRNA expression efficiency in vitro and in vivo [20].
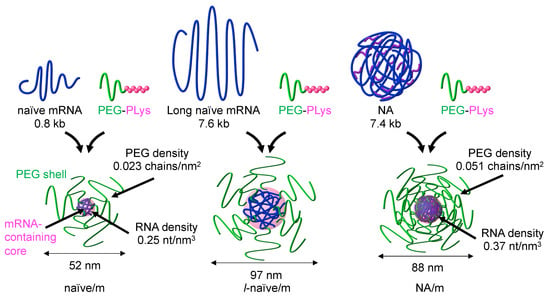
Figure 2.
Polyplex micelles (PMs). PMs were prepared by mixing mRNA and PEG-PLys block copolymer at N/P = 2 in aqueous solution and had a core–shell structure of PEG shell and mRNA-containing core. PM from naïve mRNA, that from long naïve mRNA, and that from NA are designated as naïve/m, l-naïve/m, and NA/m, respectively. Cumulant diameter was determined by DLS. Further characterization was performed for determining the density of PEG and RNA for naïve/m and NA/m. Reproduced from reference [20] with permission.
2. Methods
NAs were prepared first from Gaussia luciferase (gLuc) mRNA (783 nt) and 8 RNA oligonucleotide linkers. The hybridization of these two components was performed by heating at 65 °C for 5 min, followed by cooling to 30 °C for in 90 min. PEG-polylysine (PEG-PLys) block copolymer with PEG Mw of 12 kDa and a polymerization degree of the PLys segment of 61 was used. Note that PLys was widely used in the delivery of nucleic acids [28]. NAs and the block copolymers were mixed at (amino groups in the block copolymers (N))/(phosphate groups in NAs (P)) ratio of 2. As a control, PMs from naïve GLuc mRNA and naïve long mRNA with 7569 nt were prepared at N/P ratio of 2 and were designated as naïve/m and l-naïve/m, respectively. For examining the versatility of our strategy, NAs were also prepared from firefly luciferase (fLuc) mRNA with the length of 1854 nt, using 10 NA oligonucleotide linkers, and fLuc NA/m was prepared in the same manner as NA/m from gLuc mRNA.
3. Results and Discussion
PMs were prepared from gLuc naïve mRNA (0.8 kb) and gLuc NAs and designated as naïve/m and NA/m, respectively. To discriminate the effect of the difference in the mRNA structure from the increase in mRNA amount in NA/m compared to naïve/m, we prepared an additional control PM prepared from naïve long mRNA (l-naïve/m) (Figure 2). The length of long mRNA was set to 7.6 kb, which is close to total RNA amount in a single NA (7.4 kb), determined in our previous report [25]. All PM formulations exhibited the average sizes below 100 nm with polydispersity index around 0.15 in dynamic light scattering (DLS) analyses. Further characterization was performed to measure the molecular weight of PMs using analytical ultracentrifugation, the diameter of PM cores using transmission electron microscopic (TEM) images, and the binding ratio of block copolymer to mRNA after separating free block copolymer from PMs by ultracentrifugation. Using these values, we calculated the PEG density in the PM shell and the RNA density in the PM core. Notably, NA/m showed 1.5-fold increase in RNA density and 2.2-fold increase in PEG density (Figure 2). The compact structure of NA may be preserved after complexation with polycation, and such condensed RNA in NA/m may attract a large amount of block copolymer to increase PEG density.
Then, the physical stability of PMs was evaluated by observing RNA release from PMs after the addition of dextran sulfate, a polyanion. Such a polyion exchange reaction is a major mechanism of polyion complex dissociation in blood circulation [29,30]. NA/m required around a 1.5-fold larger amount of dextran sulfate for RNA release compared to naïve/m and l-naïve/m, demonstrating its enhanced physical stability. The biological stability of PMs was assessed by evaluating mRNA intactness after serum incubation using quantitative real-time PCR (qRT-PCR). After 15 min incubation in 50% serum, more than 60% of mRNA in NA/m was detected by qRT-PCR, while detected mRNA percentages were below 5% in naïve/m and l-naïve/m. The in vivo stability of PM was then evaluated by measuring the mRNA amount in the blood circulation using qRT-PCR, 2.5 min after intravenous injection to mice. Detected mRNA in NA/m in the blood was more than 10-fold larger than that that in naïve/m and l-naïve/m. These results demonstrate the enhanced biological stability of PMs after loading NA instead of naïve mRNA. Notably, the physical and biological stability was not improved just by elongating mRNA, as l-naïve/m failed to show a difference in stability compared to naïve/m. Thus, the enhanced stability of NA/m may be attributed to the change in mRNA structure, rather than the total size of RNA. It is reasonable to assume that the increase in the density of PEG in the shell and RNA in the core contributes to the enhanced stability.
The mRNA introduction efficiency of the PMs was evaluated first by using cultured cells. Naïve/m and NA/m showed comparable efficiency of gLuc expression when culture medium contained 10% serum. In contrast, after mRNA introduction in culture medium containing 50% serum, NA/m yielded a more than 20-fold higher level of gLuc expression compared to naïve/m. NA/m maintained its mRNA introduction efficiency in the culture medium containing a high amount of nucleases and molecules that destabilize PMs. While gLuc mRNA was used so far, we used fLuc mRNA, which is 2.4-fold longer than gLuc mRNA, to test the versatility of our approach. NA/m with an average size of 92 nm in DLS analysis was successfully prepared from fLuc mRNA and PEG-PLys block copolymer. As is the case in gLuc mRNA, fLuc NA exhibited improved physical and biological stability and mRNA introduction efficiency compared to fluc naïve mRNA. Ultimately, NA/m induced efficiency in vivo mRNA introduction in mouse brain after injection to cerebrospinal fluids.
4. Conclusions
We successfully improved the physical and biological stability of mRNA nanocarriers using an RNA nanotechnology-based approach by focusing on the mRNA steric structure. The structuring of mRNA before complexation with PEG-polycation block copolymer led to more condensed packaging of mRNA in the polyplex micelles surrounded by a denser PEG shell, which contributed to their enhanced stability. Importantly, our approach improved the functionalities of existing mRNA nanocarriers without the use of additional material, which is important for future clinical translation.
Institutional Review Board Statement
Animal experiments were conducted in accordance with the approval of the Animal Care and Use Committee of the Innovation Center of NanoMedicine, Kawasaki Institute of Industrial Promotion (Kanagawa, Japan, A16-004, 13 June 2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request.
References
- Berraondo, P.; Martini, P.G.V.; Avila, M.A.; Fontanellas, A. Messenger RNA therapy for rare genetic metabolic diseases. Gut 2019, 68, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Oehm, P.; Derhovanessian, E.; Jabulowsky, R.A.; Vormehr, M.; Gold, M.; Maurus, D.; Schwarck-Kokarakis, D.; Kuhn, A.N.; Omokoko, T.; et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 2020, 585, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Takahashi, T.; Konishi, M.; Takata, N.; Gomi, M.; Shirane, D.; Miyama, R.; Hagiwara, S.; Yamasaki, Y.; Sakurai, Y.; et al. Self-Degradable Lipid-Like Materials Based on “Hydrolysis accelerated by the intra-Particle Enrichment of Reactant (HyPER)” for Messenger RNA Delivery. Adv. Funct. Mater. 2020. [Google Scholar] [CrossRef]
- Matsui, A.; Uchida, S.; Ishii, T.; Itaka, K.; Kataoka, K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci. Rep. 2015, 5, 15810. [Google Scholar] [CrossRef]
- Crowley, S.T.; Fukushima, Y.; Uchida, S.; Kataoka, K.; Itaka, K. Enhancement of Motor Function Recovery after Spinal Cord Injury in Mice by Delivery of Brain-Derived Neurotrophic Factor mRNA. Mol. Ther. Nucleic Acids 2019, 17, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Yen, A.; Cheng, Y.; Sylvestre, M.; Gustafson, H.H.; Puri, S.; Pun, S.H. Serum Nuclease Susceptibility of mRNA Cargo in Condensed Polyplexes. Mol. Pharm. 2018, 15, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Kataoka, K. Design concepts of polyplex micelles for in vivo therapeutic delivery of plasmid DNA and messenger RNA. J. Biomed. Mater. Res. A 2019, 107, 978–990. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Zhong, Z.; Mc Cafferty, S.; Combes, F.; Huysmans, H.; De Temmerman, J.; Gitsels, A.; Vanrompay, D.; Portela Catani, J.; Sanders, N.N. mRNA therapeutics deliver a hopeful message. Nano Today 2018, 23, 16–39. [Google Scholar] [CrossRef]
- Uchida, S.; Perche, F.; Pichon, C.; Cabral, H. Nanomedicine-Based Approaches for mRNA Delivery. Mol. Pharm. 2020, 17, 3654–3684. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.J.; Brown, C.R.; Shaikh, S.; Trapp, C.; Schlegel, M.K.; Qian, K.; Sehgal, A.; Rajeev, K.G.; Jadhav, V.; Manoharan, M.; et al. Advanced siRNA Designs Further Improve In Vivo Performance of GalNAc-siRNA Conjugates. Mol. Ther. 2018, 26, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Kormann, M.S.; Hasenpusch, G.; Aneja, M.K.; Nica, G.; Flemmer, A.W.; Herber-Jonat, S.; Huppmann, M.; Mays, L.E.; Illenyi, M.; Schams, A.; et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011, 29, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Kataoka, K.; Itaka, K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics 2015, 7, 137–151. [Google Scholar] [CrossRef]
- Kawaguchi, D.; Kodama, A.; Abe, N.; Takebuchi, K.; Hashiya, F.; Tomoike, F.; Nakamoto, K.; Kimura, Y.; Shimizu, Y.; Abe, H. Phosphorothioate Modification of mRNA Accelerates the Rate of Translation Initiation to Provide More Efficient Protein Synthesis. Angew. Chem. Int. Ed. Engl. 2020. [Google Scholar] [CrossRef]
- Li, B.; Luo, X.; Dong, Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016, 27, 849–853. [Google Scholar] [CrossRef]
- Yoshinaga, N.; Uchida, S.; Naito, M.; Osada, K.; Cabral, H.; Kataoka, K. Induced packaging of mRNA into polyplex micelles by regulated hybridization with a small number of cholesteryl RNA oligonucleotides directed enhanced in vivo transfection. Biomaterials 2019, 197, 255–267. [Google Scholar] [CrossRef]
- Kurimoto, S.; Yoshinaga, N.; Igarashi, K.; Matsumoto, Y.; Cabral, H.; Uchida, S. PEG-OligoRNA Hybridization of mRNA for Developing Sterically Stable Lipid Nanoparticles toward In Vivo Administration. Molecules 2019, 24, 1303. [Google Scholar] [CrossRef]
- Koji, K.; Yoshinaga, N.; Mochida, Y.; Hong, T.; Miyazaki, T.; Kataoka, K.; Osada, K.; Cabral, H.; Uchida, S. Bundling of mRNA strands inside polyion complexes improves mRNA delivery efficiency in vitro and in vivo. Biomaterials 2020, 261, 120332. [Google Scholar] [CrossRef]
- Lee, J.B.; Hong, J.; Bonner, D.K.; Poon, Z.; Hammond, P.T. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat. Mater. 2012, 11, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Park, Y.; Kim, H.; Lee, J.B. Self-assembly of free-standing RNA membranes. Nat. Commun. 2014, 5, 4367. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Li, X.; Tian, C.; Jiang, W.; Wang, G.; Mao, C. Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nat. Commun. 2014, 5, 3890. [Google Scholar] [CrossRef] [PubMed]
- Ueki, R.; Uchida, S.; Kanda, N.; Yamada, N.; Ueki, A.; Akiyama, M.; Toh, K.; Cabral, H.; Sando, S. A chemically unmodified agonistic DNA with growth factor functionality for in vivo therapeutic application. Sci. Adv. 2020, 6, eaay2801. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, N.; Cho, E.; Koji, K.; Mochida, Y.; Naito, M.; Osada, K.; Kataoka, K.; Cabral, H.; Uchida, S. Bundling mRNA Strands to Prepare Nano-Assemblies with Enhanced Stability Towards RNase for In Vivo Delivery. Angew. Chem. Int. Ed. 2019, 58, 11360–11363. [Google Scholar] [CrossRef]
- Uchida, S.; Itaka, K.; Uchida, H.; Hayakawa, K.; Ogata, T.; Ishii, T.; Fukushima, S.; Osada, K.; Kataoka, K. In vivo messenger RNA introduction into the central nervous system using polyplex nanomicelle. PLoS ONE 2013, 8, e56220. [Google Scholar] [CrossRef]
- Uchida, S.; Kinoh, H.; Ishii, T.; Matsui, A.; Tockary, T.A.; Takeda, K.M.; Uchida, H.; Osada, K.; Itaka, K.; Kataoka, K. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials 2016, 82, 221–228. [Google Scholar] [CrossRef]
- Lachelt, U.; Wagner, E. Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef]
- Burke, R.S.; Pun, S.H. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjug. Chem. 2008, 19, 693–704. [Google Scholar] [CrossRef]
- Zuckerman, J.E.; Choi, C.H.; Han, H.; Davis, M.E. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc. Natl. Acad. Sci. USA 2012, 109, 3137–3142. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).