Preliminary Characterization of a New Processive Endoglucanase from Clostridium alkalicellulosi DSM17461 †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cloning and Protein Expression
2.2. Enzyme Activity Assay
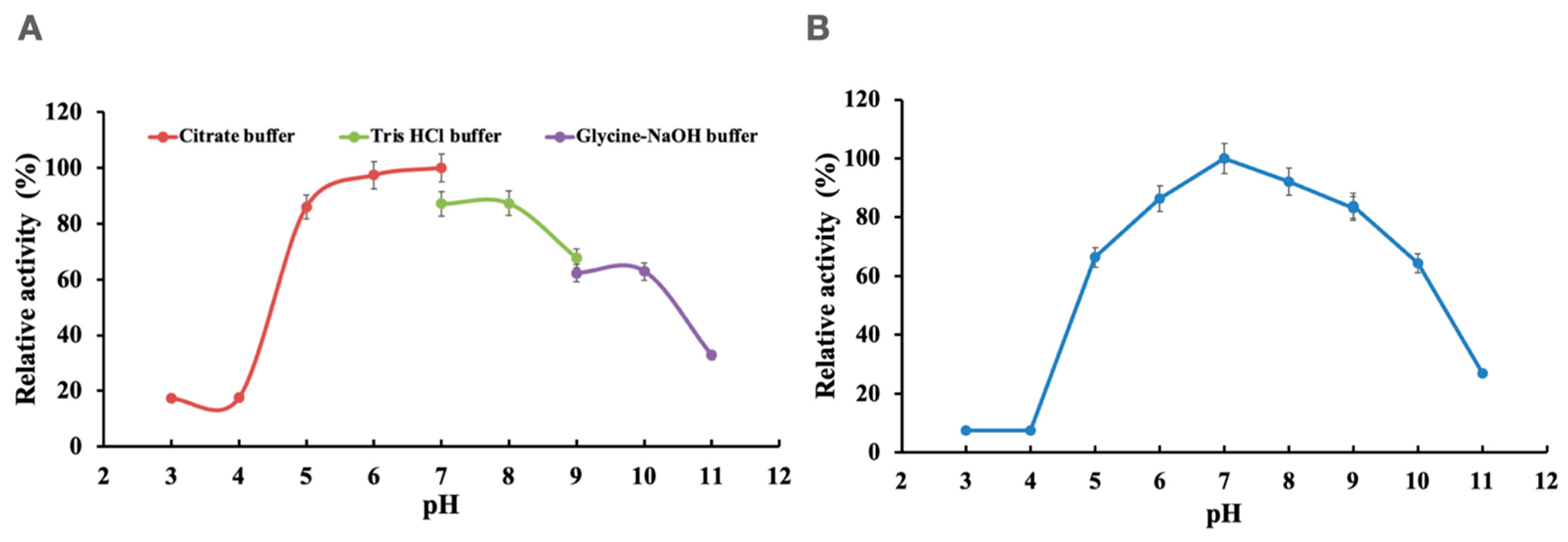
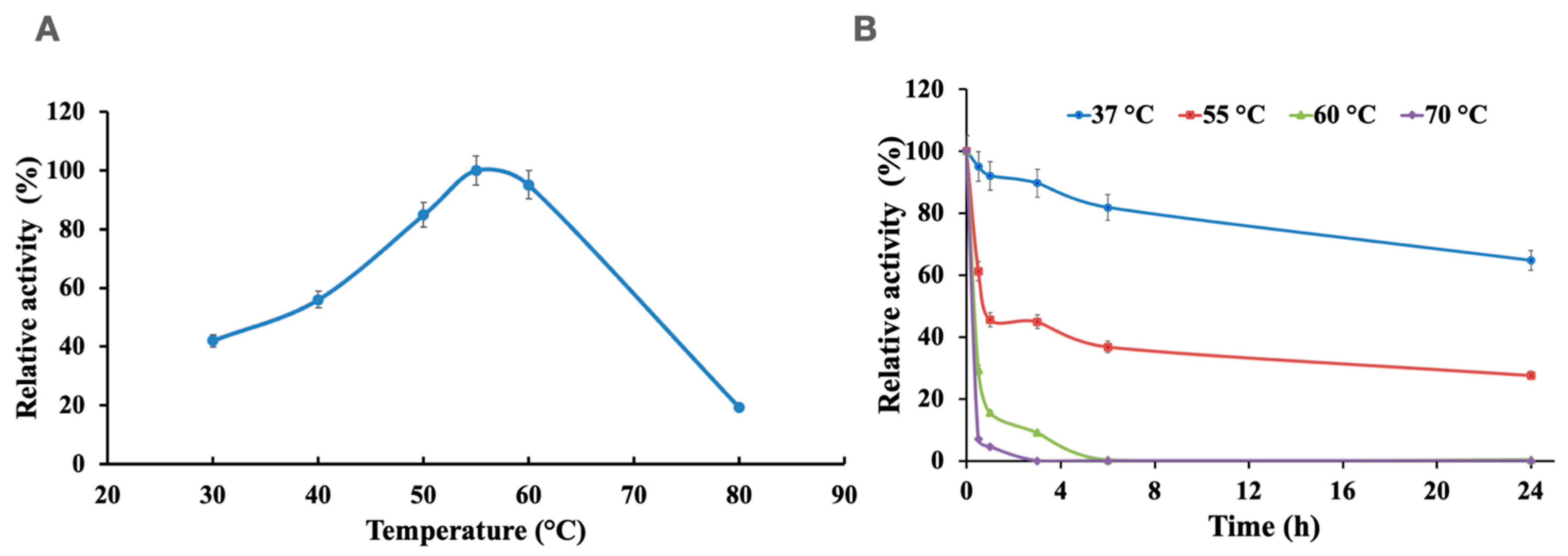
2.3. Effect of pH and Temperature
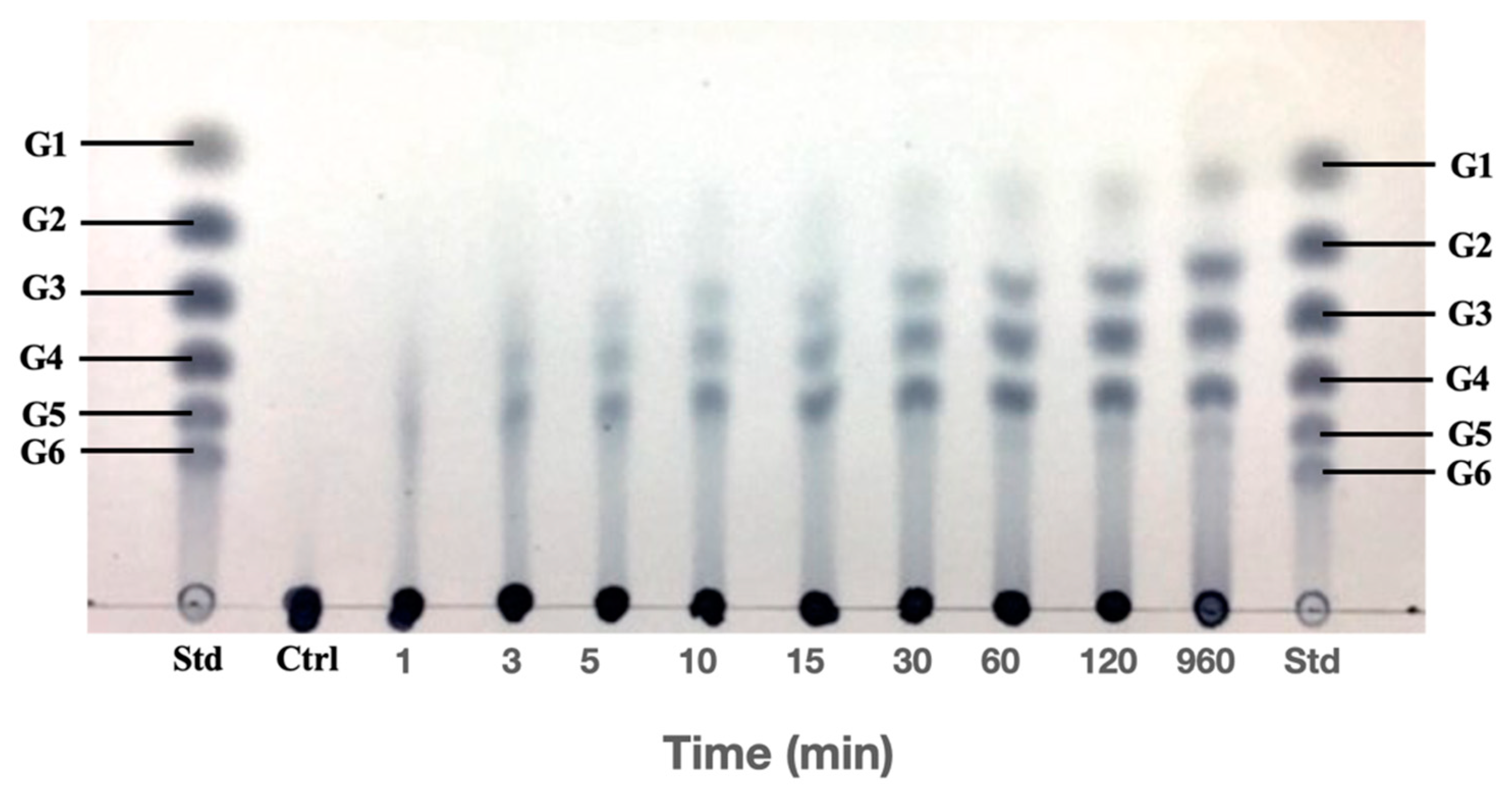
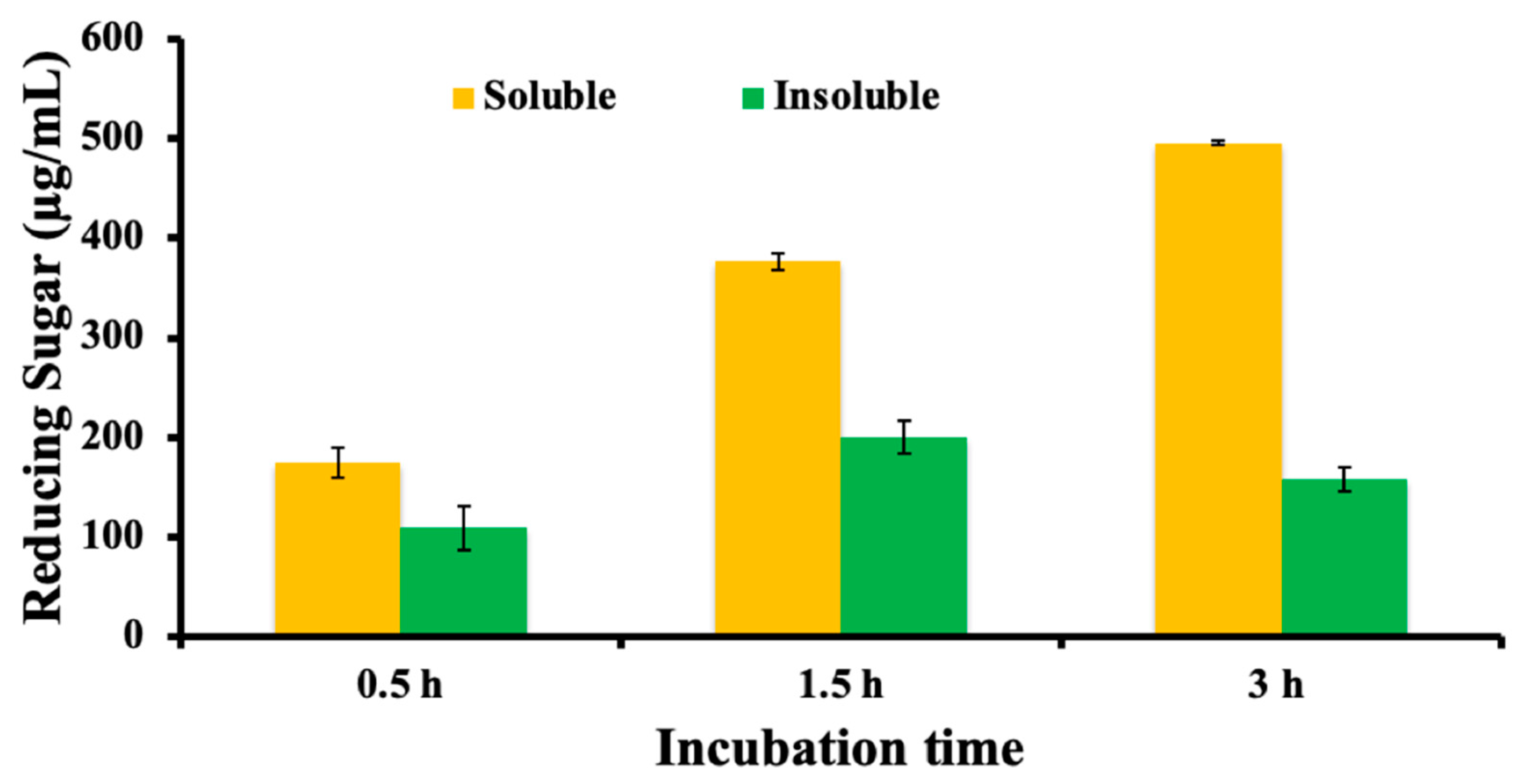
2.4. Analysis of the Hydrolysis
2.5. Processivity Assay
3. Results and Discussion
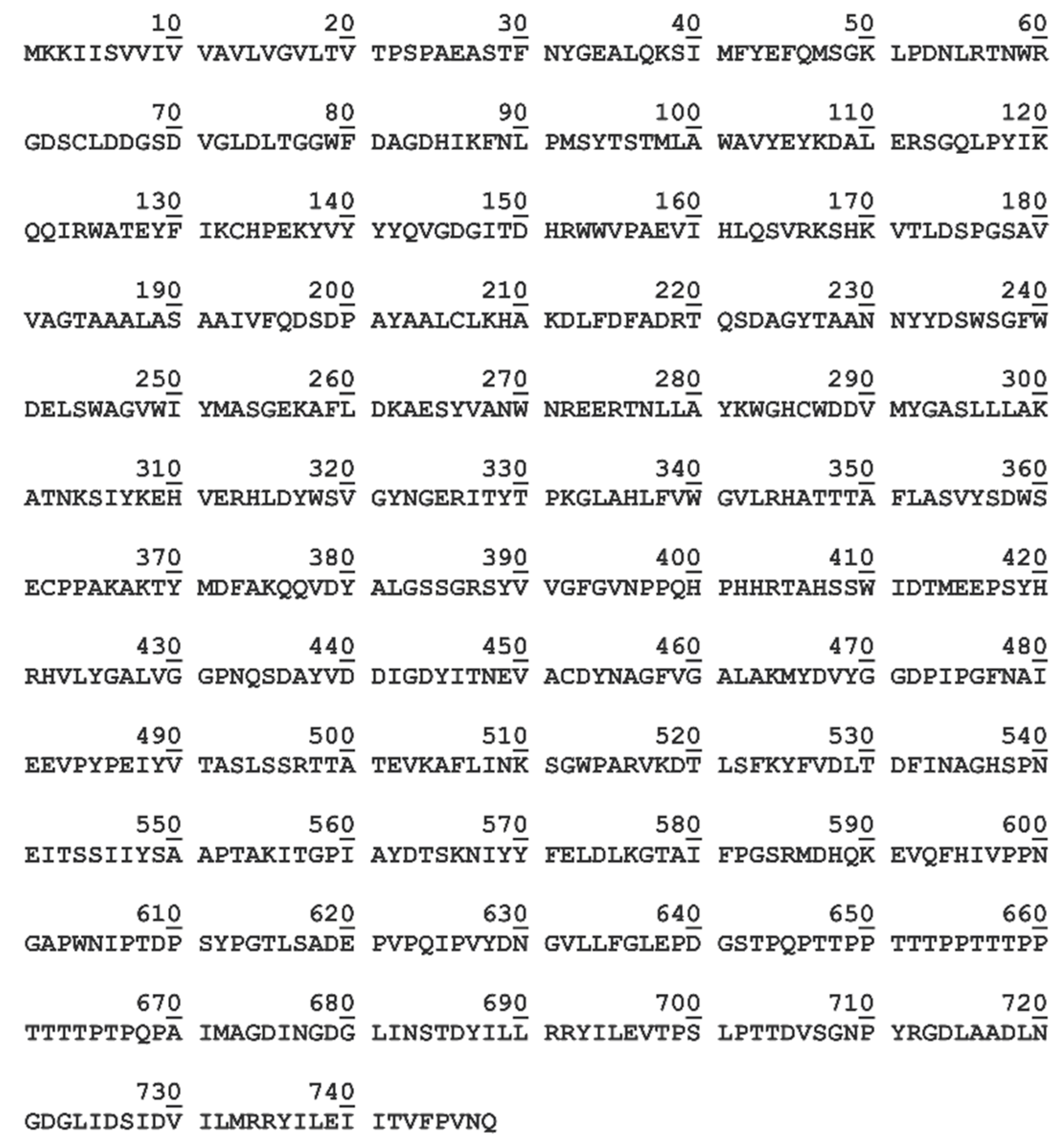
3.1. Bioinformatic Analysis
3.2. Effects of pH and Temperature
3.3. Mode of Action
3.4. Processivity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phitsuwan, P.; Sakka, K.; Ratanakhanokchai, K. Improvement of lignocellulosic biomass in planta: A review of feedstocks, biomass recalcitrance, and strategic manipulation of ideal plants designed for ethanol production and processability. Biomass Bioenergy 2013, 58, 390–405. [Google Scholar] [CrossRef]
- Himmel, M.E.; Ding, S.Y.; Johnson, D.K.; Adney, W.S.; Nimlos, M.R.; Brady, J.W.; Foust, T.D. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science 2007, 315, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, J.; Hemjinda, E.; Arai, T.; Kimura, T.; Sakka, K.; Ohmiya, K. Clostridium thermocellum cellulase CelT, a family 9 endoglucanase without an Ig-like domain or family 3c carbohydrate-binding module. Appl. Microbiol. Biotechnol. 2002, 59, 455–461. [Google Scholar] [PubMed]
- Zverlov, V.V.; Schantz, N.; Schwarz, W.H. A major new component in the cellulosome of Clostridium thermocellum is a processive endo-β-1, 4-glucanase producing cellotetraose. FEMS Microbiol. Lett. 2005, 249, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Kosugi, A.; Chan, H.; Koukiekolo, R.; Yukawa, H.; Inui, M.; Doi, R.H. Properties of cellulosomal family 9 cellulases from Clostridium cellulovorans. Appl. Microbiol. Bioiotechnol. 2006, 71, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, A.C.; Chilaka, A.C.; Church, G.M. Targeted gene inactivation in Clostridium phytofermentans shows that cellulose degradation requires the family 9 hydrolase Cphy3367. Mol. Microbiol. 2009, 74, 1300–1313. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.J.; Knox, J.P.; Boraston, A.B. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 2013, 23, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhu, Z.; Zhang, C.; Zhang, Y.-H.P. Fusion of a family 9 cellulose-binding module improves catalytic potential of Clostridium thermocellum cellodextrin phosphorylase on insoluble cellulose. Appl. Microbiol. Biotechnol. 2011, 92, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Thongekkaew, J.; Ikeda, H.; Masaki, K.; Iefuji, H. Fusion of cellulose binding domain from Trichoderma reesei CBHI to Cryptococcus sp. S-2 cellulase enhances its binding affinity and its cellulolytic activity to insoluble cellulosic substrates. Enzyme Microb. Technol. 2013, 52, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Joseph, A.; Gottumukkala, L.D. Xylanase and cellulase systems of Clostridium sp.: An insight on molecular approaches for strain improvement. Bioresour. Technol. 2014, 158, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Phitsuwan, P.; Lee, S.; San, T.; Ratanakhanokchai, K. CalkGH9T: A glycoside hydrolase family 9 enzyme from Clostridium alkalicellulosi. Catalysts 2021, 11, 1011. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Sathitsuksanoh, N.; Zhang, Y.H.P. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: Heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 2010, 101, 5534–5538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.D.; Li, W.; Wang, Y.F.; Zheng, Y.L.; Tan, F.C.; Ma, X.Q.; Yao, L.S.; Bayer, E.A.; Wang, L.S.; Li, F.L. Processive degradation of crystalline cellulose by a multimodular endoglucanase via a wirewalking mode. Biomacromolecules 2018, 19, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
San, T.; Phitsuwan, P. Preliminary Characterization of a New Processive Endoglucanase from Clostridium alkalicellulosi DSM17461. Chem. Proc. 2022, 6, 12. https://doi.org/10.3390/ECCS2021-11033
San T, Phitsuwan P. Preliminary Characterization of a New Processive Endoglucanase from Clostridium alkalicellulosi DSM17461. Chemistry Proceedings. 2022; 6(1):12. https://doi.org/10.3390/ECCS2021-11033
Chicago/Turabian StyleSan, Techly, and Paripok Phitsuwan. 2022. "Preliminary Characterization of a New Processive Endoglucanase from Clostridium alkalicellulosi DSM17461" Chemistry Proceedings 6, no. 1: 12. https://doi.org/10.3390/ECCS2021-11033
APA StyleSan, T., & Phitsuwan, P. (2022). Preliminary Characterization of a New Processive Endoglucanase from Clostridium alkalicellulosi DSM17461. Chemistry Proceedings, 6(1), 12. https://doi.org/10.3390/ECCS2021-11033






