Chemoproteomic Study of Effect of Halogenated Hydroxynaphthalenecarboxanilides on Staphylococcus aureus †
Abstract
1. Introduction
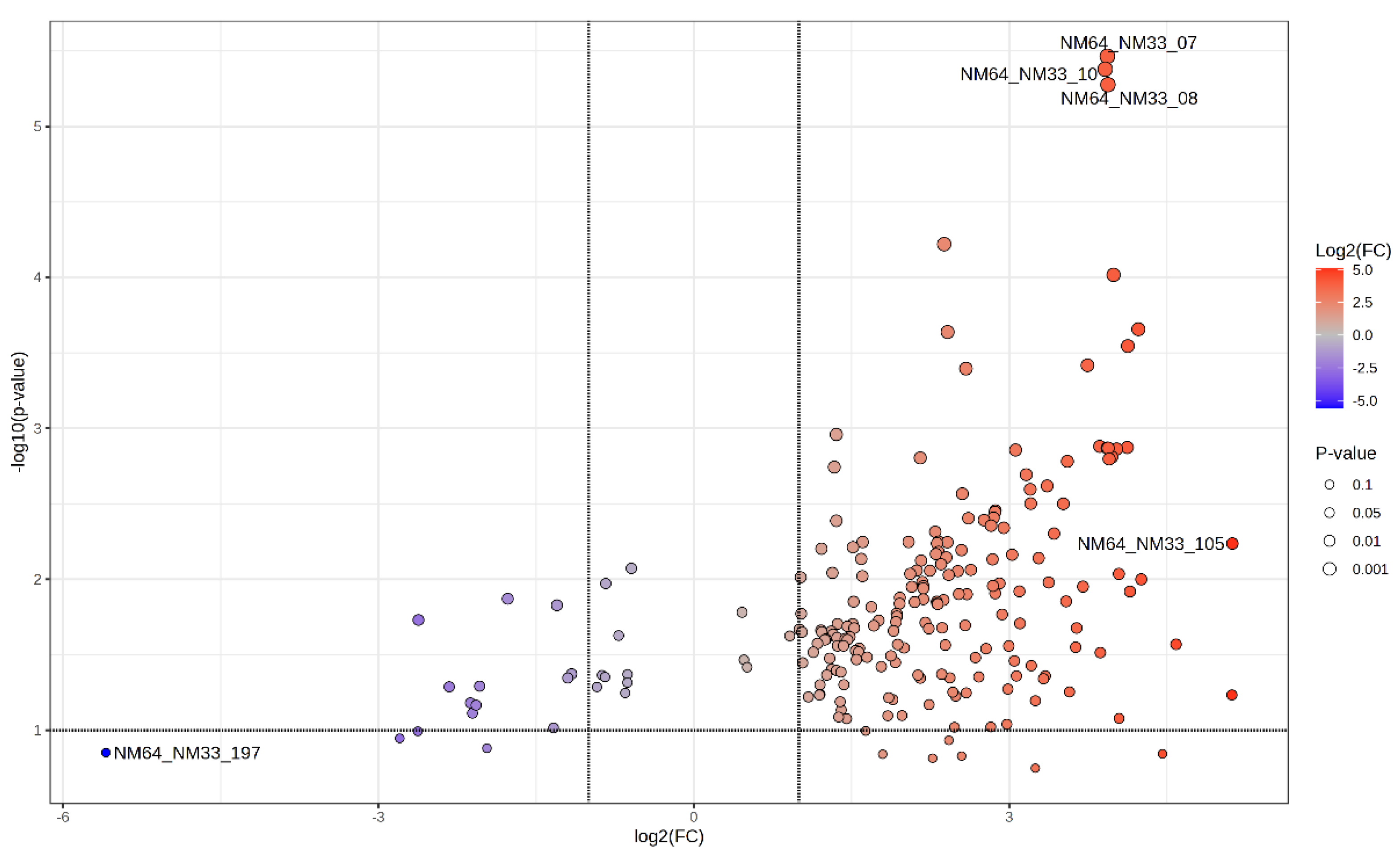
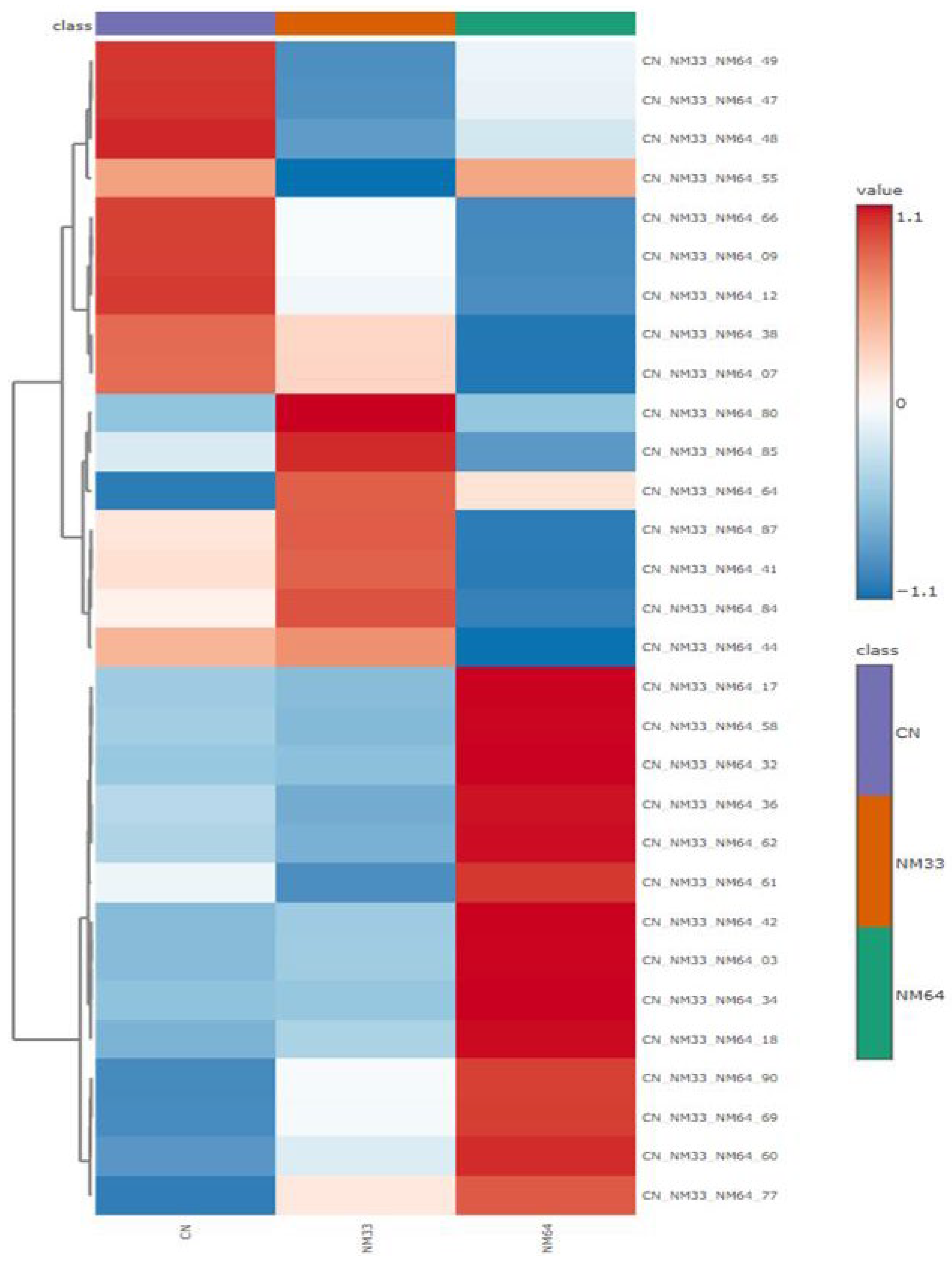
2. Results and Discussion
3. Experimental Section
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022. Available online: https://www.who.int/publications/i/item/9789240062702 (accessed on 10 September 2024).
- Suckling, C.J.; Hunter, I.S.; Scott, F.J. Multitargeted anti-infective drugs: Resilience to resistance in the antimicrobial resistance era. Future Drug Discov. 2022, 4, FDD73. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zheng, Y.; Ma, W.; Ihsan, A.; Hao, H.; Cheng, G.; Wang, X. Multitarget antibacterial drugs: An effective strategy to combat bacterial resistance. Pharmacol. Ther. 2023, 252, 108550. [Google Scholar] [CrossRef] [PubMed]
- Zadrazilova, I.; Pospisilova, S.; Masarikova, M.; Imramovsky, A.; Monreal-Ferriz, J.; Vinsova, J.; Cizek, A.; Jampilek, J. Salicylanilide carbamates: Promising antibacterial agents with high in vitro activity against methicillin-resistant Staphylococcus aureus. Eur. J. Pharm. Sci. 2015, 77, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Pindjakova, D.; Pilarova, E.; Pauk, K.; Michnova, H.; Hosek, J.; Magar, P.; Cizek, A.; Imramovsky, A.; Jampilek, J. Study of biological activities and ADMET-related properties of salicylanilide-based peptidomimetics. Int. J. Mol. Sci. 2022, 23, 11648. [Google Scholar] [CrossRef] [PubMed]
- Gonec, T.; Kos, J.; Zadrazilova, I.; Pesko, M.; Keltosova, S.; Tengler, J.; Bobal, P.; Kollar, P.; Cizek, A.; Kralova, K.; et al. Antimycobacterial and herbicidal activity of ring-substituted 1-hydroxynaphthalene-2-carboxanilides. Bioorg. Med. Chem. 2013, 21, 6531–6541. [Google Scholar] [CrossRef] [PubMed]
- Michnova, H.; Pospisilova, S.; Gonec, T.; Kapustikova, I.; Kollar, P.; Kozik, V.; Musiol, R.; Jendrzejewska, I.; Vanco, J.; Travnicek, Z.; et al. Bioactivity of methoxylated and methylated 1-hydroxynaphthalene-2-carboxanilides: Comparative molecular surface analysis. Molecules 2019, 24, 2991. [Google Scholar] [CrossRef] [PubMed]
- Gonec, T.; Pindjakova, D.; Vrablova, L.; Strharsky, T.; Michnova, H.; Kauerova, T.; Kollar, P.; Oravec, M.; Jendrzejewska, I.; Cizek, A.; et al. Antistaphylococcal activities and ADME-related properties of chlorinated arylcarbamoylnaphthalenylcarbamates. Pharmaceuticals 2022, 15, 715. [Google Scholar] [CrossRef] [PubMed]
- Spaczynska, E.; Mrozek-Wilczkiewicz, A.; Malarz, K.; Kos, J.; Gonec, T.; Oravec, M.; Gawecki, R.; Bak, A.; Dohanosova, J.; Kapustikova, I.; et al. Design and synthesis of anticancer 1-hydroxynaphthalene-2-carboxanilides with a p53 independent mechanism of action. Sci. Rep. 2019, 9, 6387. [Google Scholar] [CrossRef] [PubMed]
- Kauerova, T.; Gonec, T.; Jampilek, J.; Hafner, S.; Gaiser, A.K.; Syrovets, T.; Fedr, R.; Soucek, K.; Kollar, P. Ring-substituted 1-hydroxynaphthalene-2-carboxanilides inhibit proliferation and trigger mitochondria-mediated apoptosis. Int. J. Mol. Sci. 2020, 21, 3416. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.E.; Garibotto, F.M.; Angelina, E.; Kos, J.; Tomasic, T.; Zidar, N.; Kikelj, D.; Gonec, T.; Marvanova, P.; Mokry, P.; et al. Searching new structural scaffolds for BRAF inhibitors. An integrative study using theoretical and experimental techniques. Bioorg. Chem. 2019, 91, 103125. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.E.; Garibotto, F.; Angelina, E.; Kos, J.; Gonec, T.; Marvanova, P.; Vettorazzi, M.; Oravec, M.; Jendrzejewska, I.; Jampilek, J.; et al. Hydroxynaphthalenecarboxamides and substituted piperazinylpropandiols, two new series of BRAF inhibitors. A theoretical and experimental study. Bioorg. Chem. 2020, 103, 104145. [Google Scholar] [CrossRef] [PubMed]
- Bajorath, J. Structural characteristics of compounds with multitarget activity. Future Drug Discov. 2021, 3, FDD60. [Google Scholar] [CrossRef]
- Patil, B.S.; Chidambara Murthy, K. Bioactive compounds: Historical perspectives, opportunities, and challenges. J. Agric. Food Chem. 2009, 57, 8142–8260. [Google Scholar] [CrossRef] [PubMed]
- Moellering, R.E.; Cravatt, B.F. How chemoproteomics can enable drug discovery and development. Chem. Biol. 2012, 19, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Drewes, G.; Knapp, S. Chemoproteomics and chemical probes for target discovery. Trends Biotechnol. 2018, 36, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.H.; Neubert, H. Clinical chemoproteomics—Opportunities and obstacles. Sci. Transl. Med. 2017, 9, 7951. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wong, Y.K.; Wang, J.; Zhang, J.; Lee, Y.; Shen, H.; Lin, Q.; Hua, Z.C. Target identification with quantitative activity based protein profiling (ABPP). Proteomics 2017, 17, 1600212. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tian, Y.; Wang, M.; Wang, M.; Sun, G.; Sun, X. Advanced activity-based protein profiling application strategies for drug development. Front. Pharmacol. 2018, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.A.; Unakal, C.G. Staphylococcus aureus Infection; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK441868/ (accessed on 10 September 2024).
- UniProt: The Universal Protein Knowledgebase in 2023. Available online: https://www.uniprot.org/ (accessed on 10 September 2024).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vrablova, L.; Majerova, P.; Pindjakova, D.; Gonec, T.; Kovac, A.; Cizek, A.; Jampilek, J. Chemoproteomic Study of Effect of Halogenated Hydroxynaphthalenecarboxanilides on Staphylococcus aureus. Chem. Proc. 2024, 16, 15. https://doi.org/10.3390/ecsoc-28-20152
Vrablova L, Majerova P, Pindjakova D, Gonec T, Kovac A, Cizek A, Jampilek J. Chemoproteomic Study of Effect of Halogenated Hydroxynaphthalenecarboxanilides on Staphylococcus aureus. Chemistry Proceedings. 2024; 16(1):15. https://doi.org/10.3390/ecsoc-28-20152
Chicago/Turabian StyleVrablova, Lucia, Petra Majerova, Dominika Pindjakova, Tomas Gonec, Andrej Kovac, Alois Cizek, and Josef Jampilek. 2024. "Chemoproteomic Study of Effect of Halogenated Hydroxynaphthalenecarboxanilides on Staphylococcus aureus" Chemistry Proceedings 16, no. 1: 15. https://doi.org/10.3390/ecsoc-28-20152
APA StyleVrablova, L., Majerova, P., Pindjakova, D., Gonec, T., Kovac, A., Cizek, A., & Jampilek, J. (2024). Chemoproteomic Study of Effect of Halogenated Hydroxynaphthalenecarboxanilides on Staphylococcus aureus. Chemistry Proceedings, 16(1), 15. https://doi.org/10.3390/ecsoc-28-20152







