Potential Fluorescent Ligands for Zn-Containing Bacterial Enzymes: In Silico Evaluation, Synthesis and Optical Properties †
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Docking of CPF-Pic2, NBD-Pic2, NBD-OPD with Zn-Containing Proteins
3.2. Evaluation of Membrane Permeability In Silico
3.3. Results of Quantum Chemical Calculations for NBD-OPD Structure
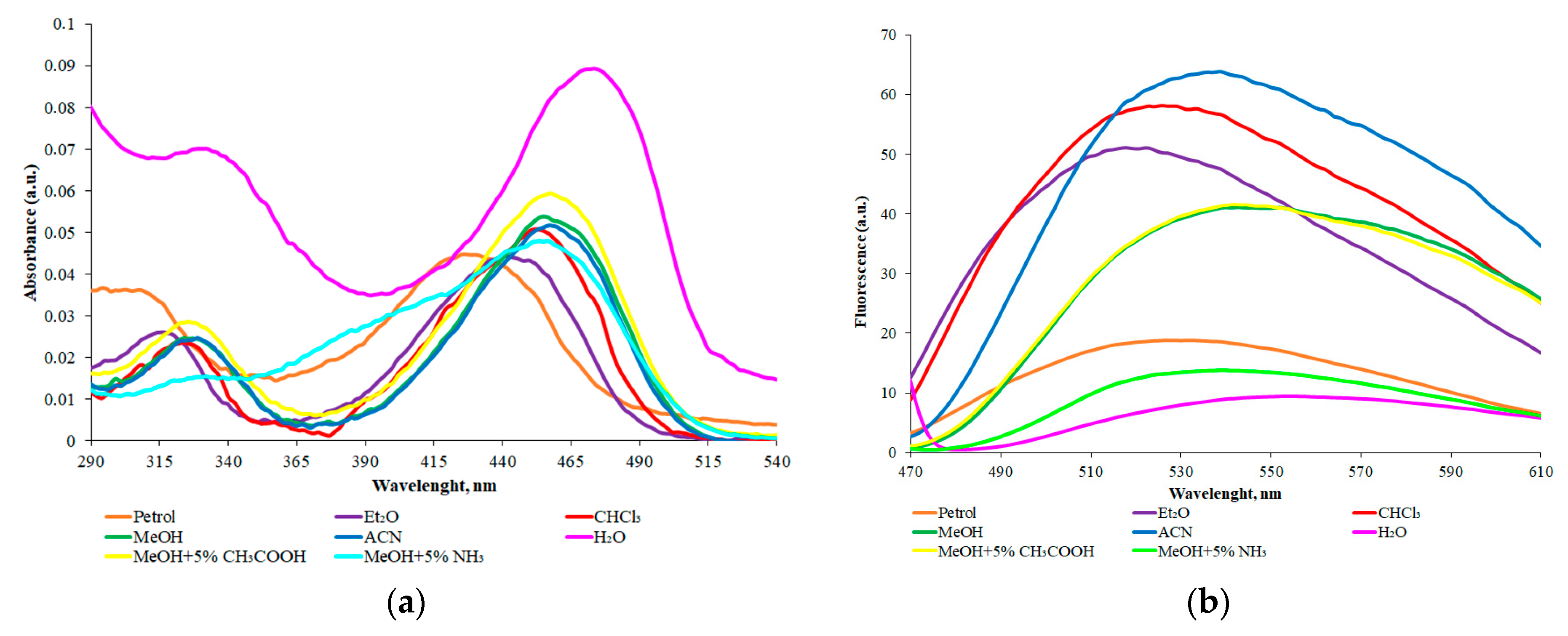
3.4. Synthesis and Solvatoshromic Properties of NBD–OPD and NBD–Pic2
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Yan, S.; Huang, R.; Feng, S.; Fu, B.; Weng, X.; Zhou, X. A turn-on fluorescent probe for detection of tyrosinase activity. J. Anal. 2013, 138, 2825–2828. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, E.S.; Torres, B.G.; Dalla Costa, T. Validation of a sensitive HPLC/fluorescence method for assessment of ciprofloxacin levels in plasma and prostate microdialysate samples from rats. Biomed. Chromatogr. 2016, 30, 330. [Google Scholar] [CrossRef] [PubMed]
- Faletrov, Y.V.; Glinskaya, L.I.; Khoretsky, M.S.; Panada, Y.V.; Frolova, N.S.; Shkumatov, V.M. Synthesis of triazole-containing ciprofloxacin conjugate and its in silico test as a cytochrome P450 ligand. J. Belarusian State Univ. Chem. 2021, 1, 21. [Google Scholar] [CrossRef]
- Gee, C.T.; Arntson, K.E.; Urick, A.K.; Mishra, N.K.; Hawk, L.M.; Wisniewski, A.J.; Pomerantz, W.C. Protein-observed 19F-NMR for fragment screening, affinity quantification and druggability assessment. Nat. Protoc. 2016, 11, 1414. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Wu, Z.Y.; Tan, H.Y.; Yan, J.W.; Liu, X.L.; Li, J.Y.; Xu, Z.Y.; Dong, C.Z.; Zhang, L. Piperazine-tuned NBD-based colorimetric and fluorescent turnoff probes for hydrogen sulfide. R. Soc. Chem. 2018, 1, 1–6. [Google Scholar]
- Hu, W.-C.; Pang, J. Ultrasensitive Detection of Bacteria Using a 2D MOF Nanozyme-Amplified Electrochemical Detector. J. Anal. Chem. 2021, 93, 8544–8552. [Google Scholar] [CrossRef] [PubMed]
- Loginova, N.V.; Koval’chuk, T.V. Redox-active metal (II) complexes of sterically hindered phenolic ligands: Antibacterial activity and reduction of cytochrome c. Part II. Metal (II) complexes of o-diphenol derivatives of thioglycolic acid. J. Polyhedron 2011, 30, 2581–2591. [Google Scholar] [CrossRef]
- Lauffer, B.E.L.; Mintzer, R. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J. Biol. Chem. 2013, 288, 26926–26943. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, C.; Dong, H.; Xu, Q.; Chou, C.J.; Zhang, Y. Synthesis and biological study of class I selective HDAC inhibitors with NO releasing activity. Bioorg. Chem. 2020, 104, 2–8. [Google Scholar]
- Rudbari, H.A.; Kordestani, N.; Cuevas-Vicario, J.V.; Zhou, M.; Efferth, T.; Correia, I.; Schirmeister, T.; Barthels, F.; Enamullah, M.; Fernandes, A.R.; et al. Investigation of the influence of chirality and halogen atoms on the anticancer activity of enantiopure palladium(II) complexes derived from chiral amino-alcohol Schiff bases and 2-picolylamine. New J. Chem. 2022, 46, 6470. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 2010, 31, 455. [Google Scholar] [CrossRef] [PubMed]
- Faletrov, Y.V.; Staravoitava, V.A.; Dudko, A.R.; Shkumatov, V.M. Application of docking-based inverse high throughput virtual screening to found phytochemical covalent inhibitors of SARS-CoV-2 main protease, NSP12 and NSP16. Res. Sq. 2022, 1, 1–20. [Google Scholar]
- Bem, M.; Badea, F.; Draghici, C.; Caproiu, M.T.; Vasilescu, M.; Voicescu, M.; Beteringhe, A.; Caragheorgheopol, A.; Maganu, M.; Constantinescu, T.; et al. Synthesis and fluorescent properties of new derivatives of 4-amino-7-nitrobenzofurazan. ARKIVOC 2007, xiii, 87–104. [Google Scholar] [CrossRef]
- Faletrov, Y.V.; Karpushenkova, V.S.; Zavalinich, V.A.; Yakovets, P.S.; Shkredava, A.D.; Shkumatov, V.M. Interaction of nitrobenzoxadiazole derivatives of piperazine and aniline with bovine serum albumine in silico and in vitro. J. Belarusian State Univ. Chem. 2021, 2, 25–35. [Google Scholar] [CrossRef]
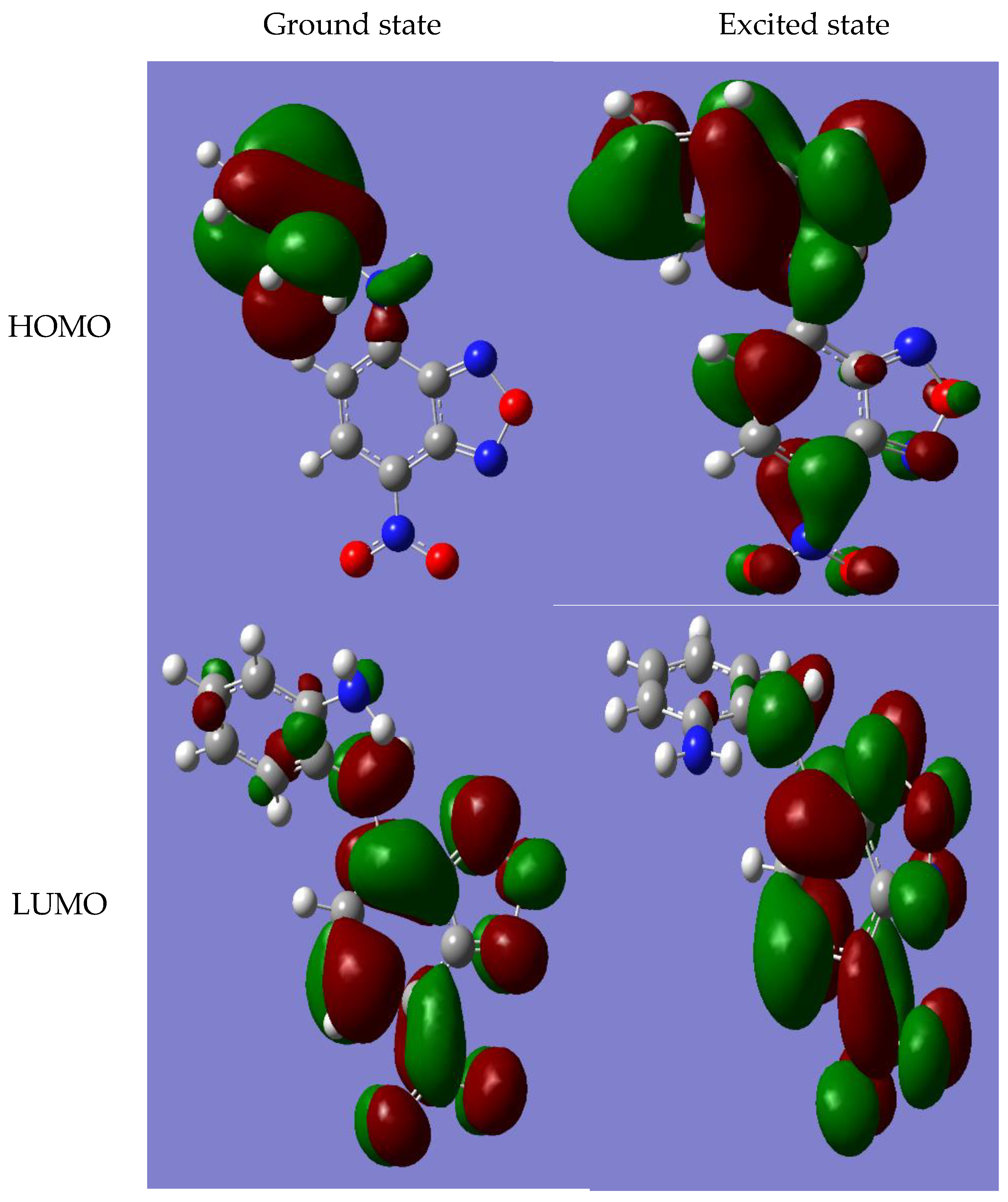
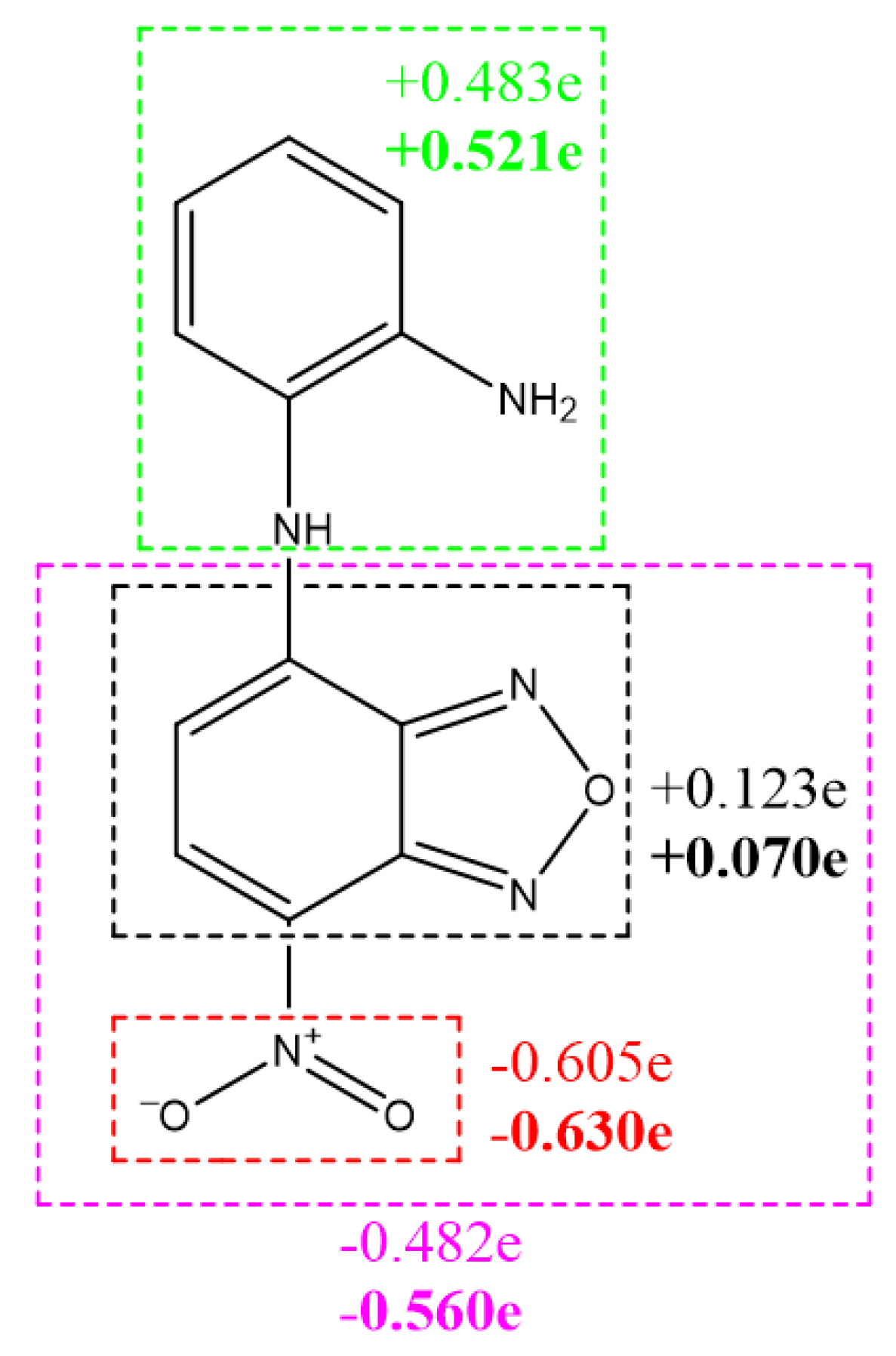
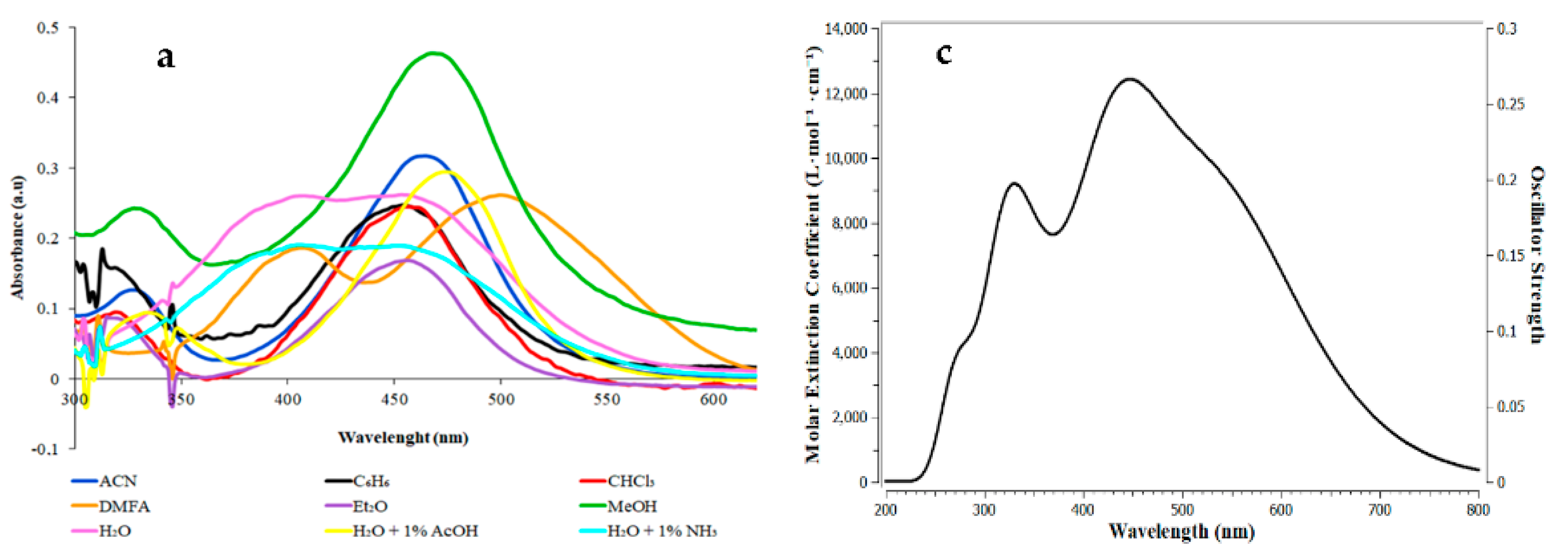
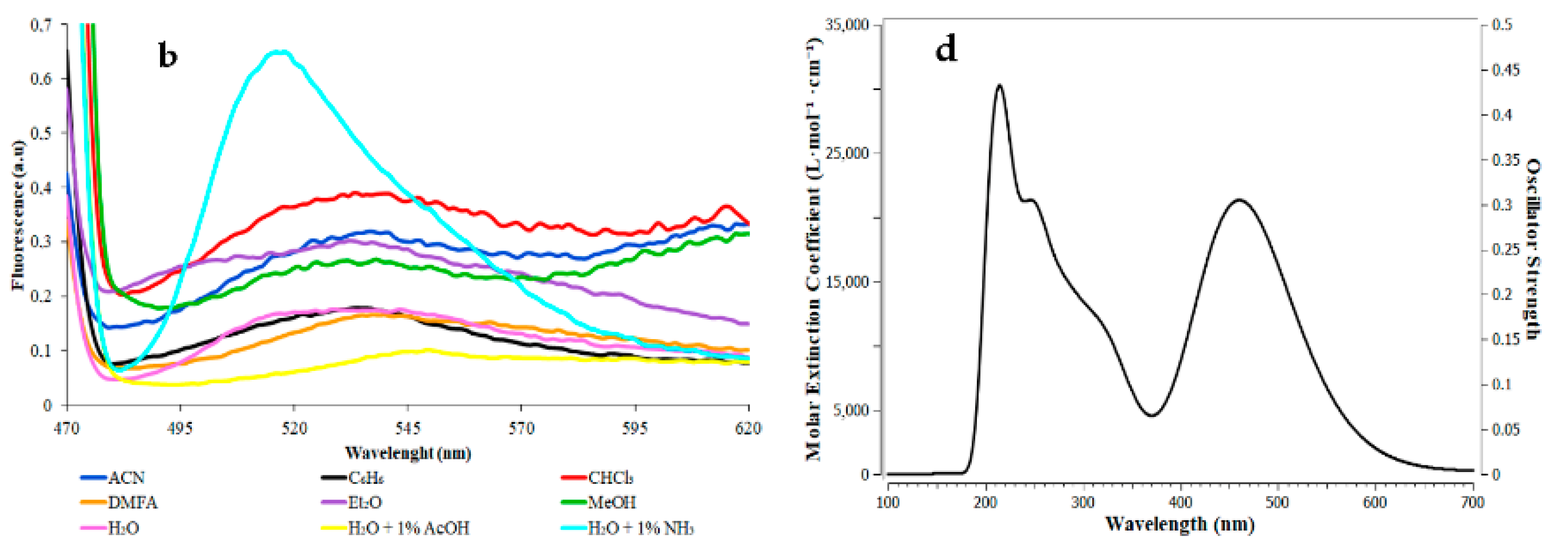
| PDB Code | Ebind | Organism | Protein | Zn- Match |
|---|---|---|---|---|
| 3HQ2 | −10 | Bacillus subtilis | M32 carboxypeptidase | + |
| 2IGI | −9.1 | Escherichia coli K-12 | Oligoribonuclease | ++ |
| 2OOG | −8.6 | Staphylococcus aureus | Phosphodiesterase | + |
| 2NQJ | −8.6 | Escherichia coli | endonuclease IV E261Q | ++ |
| 2Z29 | −8.2 | Escherichia coli | Dihydroorotase Thr109Ala | + |
| 1YT3 | −8.2 | Escherichia coli | RNAse D | ++ |
| 3GRI | −7.8 | Staphylococcus aureus | Dihydroorotase | + |
| 1Z3A | −7.7 | Escherichia coli | tRNA adenosine deaminase TadA | + |
| PDB Code | Ebind | Organism | Protein | Zn- Match |
|---|---|---|---|---|
| 2P50 | −8.9 | Escherichia coli K-12 | N-acetylglucosamine-6-phosphate deacetylase | + |
| 3ELF | −8.8 | Mycobacterium tuberculosis | Fructose-bisphosphate aldolase | + |
| 1XAH | −8.3 | Staphylococcus aureus | 3-dehydroquinate synthase | + |
| 4LEF | −8.2 | Escherichia coli K-12 | Phosphotriesterase homology protein | + |
| 2UYV | −8.1 | Escherichia coli | rhamnulose-1-phosphate aldolase | + |
| 3QBE | −8.0 | Mycobacterium tuberculosis | 3-dehydroquinate synthase | + |
| 4FUA | −8.0 | Escherichia coli | L-fuculose-1-phosphate aldolase | + |
| 1S7D | −7.9 | Escherichia coli | Metal-binding Protein yodA | + |
| PDB Code | Ebind | Organism | Protein | Zn- Match |
|---|---|---|---|---|
| 4XND | −9.4 | Escherichia coli | Isoaspartyl dipeptidase | + |
| 1RRM | −9.3 | Staphylococcus aureus | 3-dehydroquinate synthase | + |
| 2DQM | −8.7 | Staphylococcus aureus | 3-dehydroquinate synthase | + |
| 4UEJ | −8.6 | Escherichia coli K-12 | Homocysteine S-methyltransferase | + |
| 4XMX | −8.5 | Escherichia coli K-12 | Aminopeptidase N | ++ |
| 1S03 | −8.5 | Escherichia coli | Lactaldehyde reductase | + |
| 2HPT | −8.5 | Escherichia coli K-12 | N-acetylglucosamine-6-phosphate deacetylase | + |
| 5MFS | −8.2 | Staphylococcus aureus | 3-dehydroquinate synthase | ++ |
| Free Binding Energy (DOPC), kcal/mol | Logarithm of the Permeability Coefficient | |||||
|---|---|---|---|---|---|---|
| (Plasma Membrane) | (BLM) | (BBB) | (CACO2) | PAMPA-DS | ||
| NBD-Cl | −9.47 | 1.27 | - | −1.52 | −2.48 | - |
| OPD | −2.31 | −3.61 | −2.14 | −3.65 | −4.03 | −3.20 |
| NBD-OPD | −4.77 | −2.71 | −1.02 | −3.26 | −3.74 | −2.17 |
| CPF-Pic2 | −2.32 | - | −5.64 | −4.88 | −4.92 | −6.43 |
| NBD-Pic2 | −4.45 | −0.58 | - | −3.10 | −3.63 | - |
| Intramolecular Short Contacts (Å) | Covalent Bonds (Å) | Dihedral Angle | Dipole Moment (D) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| D1 O···H | D2 N···O | D3 N···H | D4 N···H | D5 N···H | dC-N (1) | dC-N (2) | ϕ (°) | ||
| NBD-Cl | 2.391 2.37 | 2.797 2.756 | - | - | - | 1.452 1.428 | - | - | 7.235 7.980 |
| OPD | - | - | 2.656 2.652 | 2.649 2.657 | - | - | - | - | 3.385 3.305 |
| NBD-OPD | 2.393 2.406 | 2.807 2.782 | 2.679 2.719 | 2.670 2.657 | 2.513 2.596 | 1.420 1.390 | 1.349 1.417 | 128.63 94.778 | 14.55 12.403 |
| Compound | Energy Levels | Chemical Reactivity Descriptors | |||
|---|---|---|---|---|---|
| εHOMO (eV) | εLUMO (eV) | μ (eV) | η (eV) | W (eV) | |
| NBD-Cl | −7.621 | −3.916 | −5.769 | 1.853 | 8.981 |
| OPD | −5.469 | −0.2584 | −2.864 | 2.605 | 1.574 |
| NBD-OPD | −6.046 | −3.355 | −4.701 | 1.346 | 8.211 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zavalinich, V.; Glinskaya, L.; Yakovets, P.; Faletrov, Y.; Shkumatov, V. Potential Fluorescent Ligands for Zn-Containing Bacterial Enzymes: In Silico Evaluation, Synthesis and Optical Properties. Chem. Proc. 2022, 12, 82. https://doi.org/10.3390/ecsoc-26-13685
Zavalinich V, Glinskaya L, Yakovets P, Faletrov Y, Shkumatov V. Potential Fluorescent Ligands for Zn-Containing Bacterial Enzymes: In Silico Evaluation, Synthesis and Optical Properties. Chemistry Proceedings. 2022; 12(1):82. https://doi.org/10.3390/ecsoc-26-13685
Chicago/Turabian StyleZavalinich, Viktoryia, Liliya Glinskaya, Polina Yakovets, Yaroslav Faletrov, and Vladimir Shkumatov. 2022. "Potential Fluorescent Ligands for Zn-Containing Bacterial Enzymes: In Silico Evaluation, Synthesis and Optical Properties" Chemistry Proceedings 12, no. 1: 82. https://doi.org/10.3390/ecsoc-26-13685
APA StyleZavalinich, V., Glinskaya, L., Yakovets, P., Faletrov, Y., & Shkumatov, V. (2022). Potential Fluorescent Ligands for Zn-Containing Bacterial Enzymes: In Silico Evaluation, Synthesis and Optical Properties. Chemistry Proceedings, 12(1), 82. https://doi.org/10.3390/ecsoc-26-13685





