Preparation and Hydro-Lipophilic Properties of Monosubstituted N-Aryl-4-hydroxyquinoline-3-carboxanilides †
Abstract
1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. General Methods
3.2. Synthesis
General Procedure for Synthesis of Carboxamides 1–8c
3.3. Lipophilicity Determination by HPLC
3.4. Lipophilicity Calculations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kruger, A.; Maltarollo, V.G.; Wrenger, C.; Kronenberger, T. ADME profiling in drug discovery and a new path paved on silica. In Drug Discovery and Development—New Advances; Gaitonde, V., Karmakar, P., Trivedi, A., Eds.; IntechOpen: Rijeka, Croatia, 2019; Available online: https://www.intechopen.com/chapters/66969 (accessed on 4 October 2022).
- Roy, A. Early probe and drug discovery in academia: A minireview. High-Throughput 2018, 7, 4. [Google Scholar] [CrossRef]
- Chmiel, T.; Mieszkowska, A.; Kempinska-Kupczyk, D.; Kot-Wasik, A.; Namiesnik, J.; Mazerska, Z. The impact of lipophilicity on environmental processes, drug delivery and bioavailability of food components. Microchem. J. 2019, 146, 393–406. [Google Scholar] [CrossRef]
- Pyka-Pająk, A.; Parys, W.; Dołowy, M. Comparison of the utility of RP-TLC technique and different computational methods to assess the lipophilicity of selected antiparasitic, antihypertensive, and anti-inflammatory drugs. Molecules 2019, 24, 3187. [Google Scholar] [CrossRef]
- Kerns, E.H.; Di, L. Drug-Like Properties: Concepts, Structure Design and Methods: From ADME to Toxicity Optimization; Academic Press: San Diego, CA, USA, 2008. [Google Scholar]
- Rutkowska, E.; Pajak, K.; Jozwiak, K. Lipophilicity—Methods of determination and its role in medicinal chemistry. Acta Pol. Pharm. 2013, 70, 3–18. [Google Scholar]
- Pliska, V.; Testa, B.; van der Waterbeemd, H. Lipophilicity in Drug Action and Toxicology; Wiley-VCH: Weinheim, Germany, 1996. [Google Scholar]
- Soares, J.X.; Santos, A.; Fernandes, C.; Pinto, M.M.M. Liquid chromatography on the different methods for the determination of lipophilicity: An essential analytical tool in medicinal chemistry. Chemosensors 2022, 10, 340. [Google Scholar] [CrossRef]
- Kempinska, D.; Chmiel, T.; Kot-Wasik, A.; Mroz, A.; Mazerska, Z.; Namiesnik, J. State of the art and prospects of methods for determination of lipophilicity of chemical compounds. Trends Anal. Chem. 2019, 113, 54–73. [Google Scholar] [CrossRef]
- Jampilek, J.; Dolowy, M.; Pyka-Pajak, A. Estimating limits of detection and quantification of ibuprofen by TLC-densitometry at different chromatographic conditions. Processes 2020, 8, 919. [Google Scholar] [CrossRef]
- Polanski, J.; Kurczyk, A.; Bak, A.; Musiol, R. Privileged structures—Dream or reality: Preferential organization of azanaphthalene scaffold. Curr. Med. Chem. 2012, 19, 1921–1945. [Google Scholar] [CrossRef]
- Jampilek, J. Recent advances in design of potential quinoxaline anti-infectives. Curr. Med. Chem. 2014, 21, 4347–4373. [Google Scholar] [CrossRef]
- Singh, S.; Kaur, G.; Mangla, V.; Gupta, M.K. Quinoline and quinolones: Promising scaffolds for future antimycobacterial agents. J. Enzyme Inhib. Med. Chem. 2015, 30, 492–504. [Google Scholar] [CrossRef]
- Steinhilber, D.; Schubert-Zsilavecz, M.; Roth, H.J. Infektionen–Antiprotozoische Wirkstoffe. In Medizinische Chemie: Targets, Arzneistoffe, Chemische Biologie; Deutscher Apotheker Verlag: Stutgart, Germany, 2010; pp. 586–593. [Google Scholar]
- Shah, S.; Dalecki, A.G.; Malalasekera, A.P.; Crawford, C.L.; Michalek, S.M.; Kutsch, O.; Sun, J.; Bossmann, S.H.; Wolschendorf, F. 8-Hydroxyquinolines are boosting agents of copper-related toxicity in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2016, 60, 5765–5776. [Google Scholar] [CrossRef]
- Mathew, B.; Ross, L.; Reynolds, R.C. A novel quinoline derivative that inhibits mycobacterial FtsZ. Tuberculosis 2013, 93, 398–400. [Google Scholar] [CrossRef]
- Paritala, H.; Carroll, K.S. New targets and inhibitors of mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 2013, 13, 85–115. [Google Scholar] [CrossRef]
- Khan, S.R.; Singh, S.; Roy, K.K.; Akhtar, M.S.; Saxena, A.K.; Krishnan, M.Y. Biological evaluation of novel substituted chloroquinolines targeting mycobacterial ATP synthase. Int. J. Antimicrob. Agents 2013, 41, 41–46. [Google Scholar] [CrossRef]
- Naik, M.; Humnabadkar, V.; Tantry, S.J.; Panda, M.; Narayan, A.; Guptha, S.; Panduga, V.; Manjrekar, P.; Jena, L.K.; Koushik, K.; et al. 4-Aminoquinolone piperidine amides: Noncovalent inhibitors of DprE1 with long residence time and potent antimycobacterial activity. J. Med. Chem. 2014, 57, 5419–5434. [Google Scholar] [CrossRef]
- Bueno, R.V.; Braga, R.C.; Segretti, N.D.; Ferreira, E.I.; Trossini, G.H.; Andrade, C.H. New tuberculostatic agents targeting nucleic acid biosynthesis: Drug design using QSAR approaches. Curr. Pharm. Des. 2014, 20, 4474–4485. [Google Scholar] [CrossRef]
- Aldred, K.J.; Blower, T.R.; Kerns, R.J.; Berger, J.M.; Osheroff, N. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proc. Natl. Acad. Sci. USA 2016, 113, 839–846. [Google Scholar] [CrossRef]
- Kuhn, F.M.L.; Alexander, E.; Minasov, G.; Page, H.J.; Warwrzak, Z.; Shuvalova, L.; Flores, K.J.; Wilson, D.J.; Shi, C.; Aldrich, C.C.; et al. Structure of the essential Mtb FadD32 enzyme: A promising drug target for treating tuberculosis. ACS Infect. Dis. 2016, 2, 579–591. [Google Scholar] [CrossRef]
- Fernandes, G.F.D.S.; Man-Chin, C.; Dos Santos, J.L. Advances in drug discovery of new antitubercular multidrug-resistant compounds. Pharmaceuticals 2017, 10, 51. [Google Scholar] [CrossRef]
- Musiol, R. Styrylquinoline—A versatile scaffold in medicinal chemistry. Med. Chem. 2020, 16, 141–154. [Google Scholar] [CrossRef]
- Musiol, R.; Jampilek, J.; Podeszwa, B.; Finster, J.; Tabak, D.; Dohnal, J.; Polanski, J. RP-HPLC determination of drug lipophilicity in series of quinoline derivatives. Cent. Eur. J. Chem. 2009, 7, 586–597. [Google Scholar]
- Jampilek, J.; Musiol, R.; Finster, J.; Pesko, M.; Carroll, J.; Kralova, K.; Vejsova, M.; O’Mahony, J.; Coffey, A.; Dohnal, J.; et al. Investigating biological activity spectrum for novel styrylquinazoline analogues. Molecules 2009, 14, 4246–4265. [Google Scholar] [CrossRef]
- Musiol, R.; Jampilek, J.; Nycz, J.E.; Pesko, M.; Carroll, J.; Kralova, K.; Vejsova, M.; O’Mahony, J.; Coffey, A.; Mrozek, A.; et al. Investigating the activity spectrum for ring-substituted 8-hydroxyquinolines. Molecules 2010, 15, 288–304. [Google Scholar] [CrossRef]
- Mrozek-Wilczkiewicz, A.; Kalinowski, D.; Musiol, R.; Finster, J.; Szurko, A.; Serafin, K.; Knas, M.; Kamalapuram, S.K.; Kovacevic, Z.; Jampilek, J.; et al. Investigating anti-proliferative activity of styrylazanaphthalenes and azanaphthalenediones. Bioorg. Med. Chem. 2010, 18, 2664–2671. [Google Scholar] [CrossRef]
- Gonec, T.; Bobal, P.; Sujan, J.; Pesko, M.; Guo, J.; Kralova, K.; Pavlacka, L.; Vesely, L.; Kreckova, E.; Kos, J.; et al. Investigating the spectrum of biological activity of substituted quinoline-2-carboxamides and their isosteres. Molecules 2012, 17, 613–644. [Google Scholar] [CrossRef]
- Kos, J.; Zadrazilova, I.; Nevin, E.; Soral, M.; Gonec, T.; Kollar, P.; Oravec, M.; Coffey, A.; O’Mahony, J.; Liptaj, T.; et al. Ring-substituted 8-hydroxyquinoline-2-carboxanilides as potential antimycobacterial agents. Bioorg. Med. Chem. 2015, 23, 4188–4196. [Google Scholar] [CrossRef]
- Gonec, T.; Kos, J.; Zadrazilova, I.; Pesko, M.; Keltosova, S.; Tengler, J.; Bobal, P.; Kollar, P.; Cizek, A.; Kralova, K.; et al. Antimycobacterial and herbicidal activity of ring-substituted 1-hydroxynaphthalene-2-carboxanilides. Bioorg. Med. Chem. 2013, 21, 6531–6541. [Google Scholar] [CrossRef]
- Kos, J.; Zadrazilova, I.; Pesko, M.; Keltosova, S.; Tengler, J.; Gonec, T.; Bobal, P.; Kauerova, T.; Oravec, M.; Kollar, P.; et al. Antibacterial and herbicidal activity of ring-substituted 3-hydroxynaphthalene-2-carboxanilides. Molecules 2013, 18, 7977–7997. [Google Scholar] [CrossRef]
- Kos, J.; Nevin, E.; Soral, M.; Kushkevych, I.; Gonec, T.; Bobal, P.; Kollar, P.; Coffey, A.; O’Mahony, J.; Liptaj, T.; et al. Synthesis and antimycobacterial properties of ring-substituted 6-hydroxynaphthalene-2-carboxanilides. Bioorg. Med. Chem. 2015, 23, 2035–2043. [Google Scholar] [CrossRef]
- Pavic, K.; Perkovic, I.; Pospisilova, S.; Machado, M.; Fontinha, D.; Prudencio, M.; Jampilek, J.; Coffey, A.; Endersen, L.; Rimac, H.; et al. Primaquine hybrids as promising antimycobacterial and antimalarial agents. Eur. J. Med. Chem. 2018, 143, 769–779. [Google Scholar] [CrossRef]
- Kapustikova, I.; Bak, A.; Gonec, T.; Kos, J.; Kozik, V.; Jampilek, J. Investigation of hydro-lipophilic properties of N-alkoxyphenylhydroxynaphthalenecarboxamides. Molecules 2018, 23, 1635. [Google Scholar] [CrossRef]
- Michnova, H.; Pospisilova, S.; Gonec, T.; Kapustikova, I.; Kollar, P.; Kozik, V.; Musiol, R.; Jendrzejewska, I.; Vanco, J.; Travnicek, Z.; et al. Bioactivity of methoxylated and methylated 1-hydroxynaphthalene-2-carboxanilides: Comparative molecular surface analysis. Molecules 2019, 24, 2991. [Google Scholar] [CrossRef]
- Bak, A.; Kos, J.; Michnova, H.; Gonec, T.; Pospisilova, S.; Kozik, V.; Cizek, A.; Smolinski, A.; Jampilek, J. Consensus-based pharmacophore mapping for new set of N-(disubstituted-phenyl)-3-hydroxyl-naphthalene-2-carboxamides. Int. J. Mol. Sci. 2020, 21, 6583. [Google Scholar] [CrossRef]
- Gonec, T.; Pindjakova, D.; Vrablova, L.; Strharsky, T.; Michnova, H.; Kauerova, T.; Kollar, P.; Oravec, M.; Jendrzejewska, I.; Cizek, A.; et al. Antistaphylococcal activities and adme-related properties of chlorinated arylcarbamoylnaphthalenylcarbamates. Pharmaceuticals 2022, 15, 715. [Google Scholar] [CrossRef]


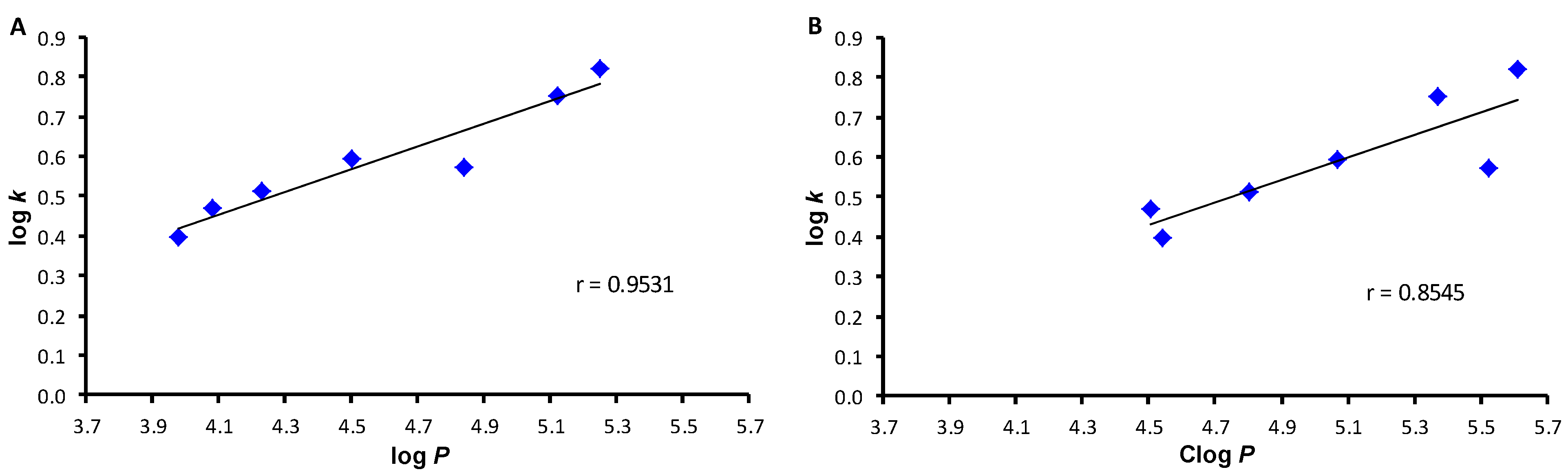

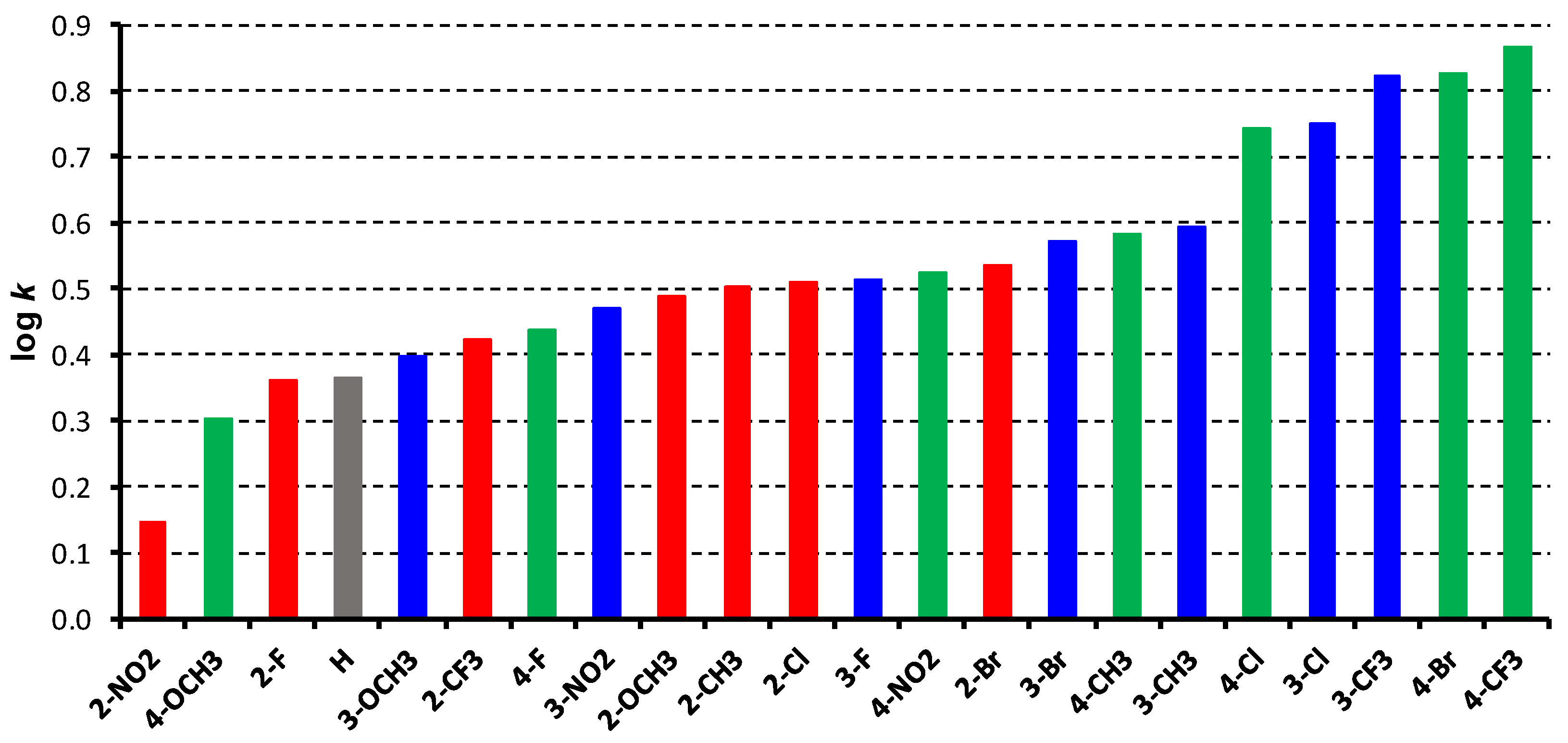
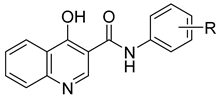 | |||||
|---|---|---|---|---|---|
| Comp. | R | log k | log P 1 | log P 2 | Clog P 2 |
| 1 | H | 0.3655 | 3.93 | 2.53 | 4.5695 |
| 2a | 2-OCH3 | 0.4873 | 4.05 | 2.41 | 3.9533 |
| 2b | 3-OCH3 | 0.3956 | 3.98 | 2.41 | 4.5433 |
| 2c | 4-OCH3 | 0.3019 | 3.80 | 2.41 | 4.5433 |
| 3a | 2-CH3 | 0.5033 | 4.50 | 3.02 | 4.4185 |
| 3b | 3-CH3 | 0.5916 | 4.50 | 3.02 | 5.0685 |
| 3c | 4-CH3 | 0.5840 | 4.50 | 3.02 | 5.0685 |
| 4a | 2-F | 0.3591 | 3.95 | 2.69 | 4.2027 |
| 4b | 3-F | 0.5126 | 4.23 | 2.69 | 4.8027 |
| 4c | 4-F | 0.4383 | 4.14 | 2.69 | 4.8027 |
| 5a | 2-Cl | 0.5086 | 4.83 | 3.09 | 4.5227 |
| 5b | 3-Cl | 0.7499 | 5.12 | 3.09 | 5.3727 |
| 5c | 4-Cl | 0.7434 | 4.93 | 3.09 | 5.3727 |
| 6a | 2-Br | 0.5347 | 4.82 | 3.36 | 4.6427 |
| 6b | 3-Br | 0.5702 | 4.84 | 3.36 | 5.5227 |
| 6c | 4-Br | 0.8269 | 4.80 | 3.36 | 5.5227 |
| 7a | 2-CF3 | 0.4228 | 5.05 | 3.45 | 4.1603 |
| 7b | 3-CF3 | 0.8211 | 5.25 | 3.45 | 5.6103 |
| 7c | 4-CF3 | 0.8672 | 5.05 | 3.45 | 5.6103 |
| 8a | 2-NO2 | 0.1446 | 4.03 | 2.40 | 4.0457 |
| 8b | 3-NO2 | 0.4697 | 4.08 | 2.40 | 4.5057 |
| 8c | 4-NO2 | 0.5238 | 3.89 | 2.40 | 4.5057 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonec, T.; Vrablova, L.; Pindjakova, D.; Strharsky, T.; Oravec, M.; Jampilek, J. Preparation and Hydro-Lipophilic Properties of Monosubstituted N-Aryl-4-hydroxyquinoline-3-carboxanilides. Chem. Proc. 2022, 12, 28. https://doi.org/10.3390/ecsoc-26-13548
Gonec T, Vrablova L, Pindjakova D, Strharsky T, Oravec M, Jampilek J. Preparation and Hydro-Lipophilic Properties of Monosubstituted N-Aryl-4-hydroxyquinoline-3-carboxanilides. Chemistry Proceedings. 2022; 12(1):28. https://doi.org/10.3390/ecsoc-26-13548
Chicago/Turabian StyleGonec, Tomas, Lucia Vrablova, Dominika Pindjakova, Tomas Strharsky, Michal Oravec, and Josef Jampilek. 2022. "Preparation and Hydro-Lipophilic Properties of Monosubstituted N-Aryl-4-hydroxyquinoline-3-carboxanilides" Chemistry Proceedings 12, no. 1: 28. https://doi.org/10.3390/ecsoc-26-13548
APA StyleGonec, T., Vrablova, L., Pindjakova, D., Strharsky, T., Oravec, M., & Jampilek, J. (2022). Preparation and Hydro-Lipophilic Properties of Monosubstituted N-Aryl-4-hydroxyquinoline-3-carboxanilides. Chemistry Proceedings, 12(1), 28. https://doi.org/10.3390/ecsoc-26-13548







