A Structure-Guided Designed Small Molecule Is an Anticancer Agent and Inhibits the Apoptosis-Related MCL-1 Protein
Abstract
:1. Introduction
2. Experimental and Computational Procedures
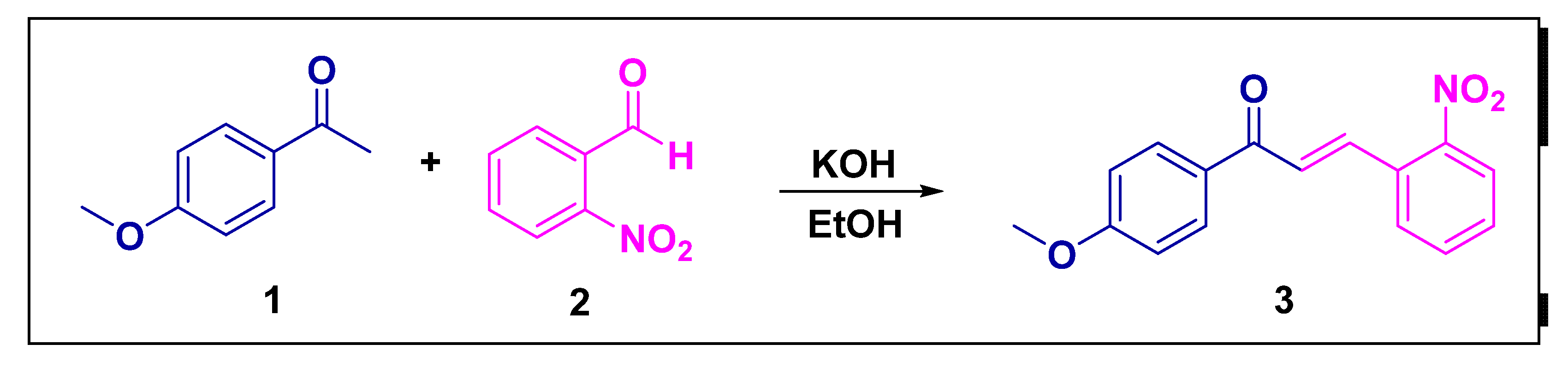
2.1. Synthesis and Crystallization
2.2. Structure Characterization
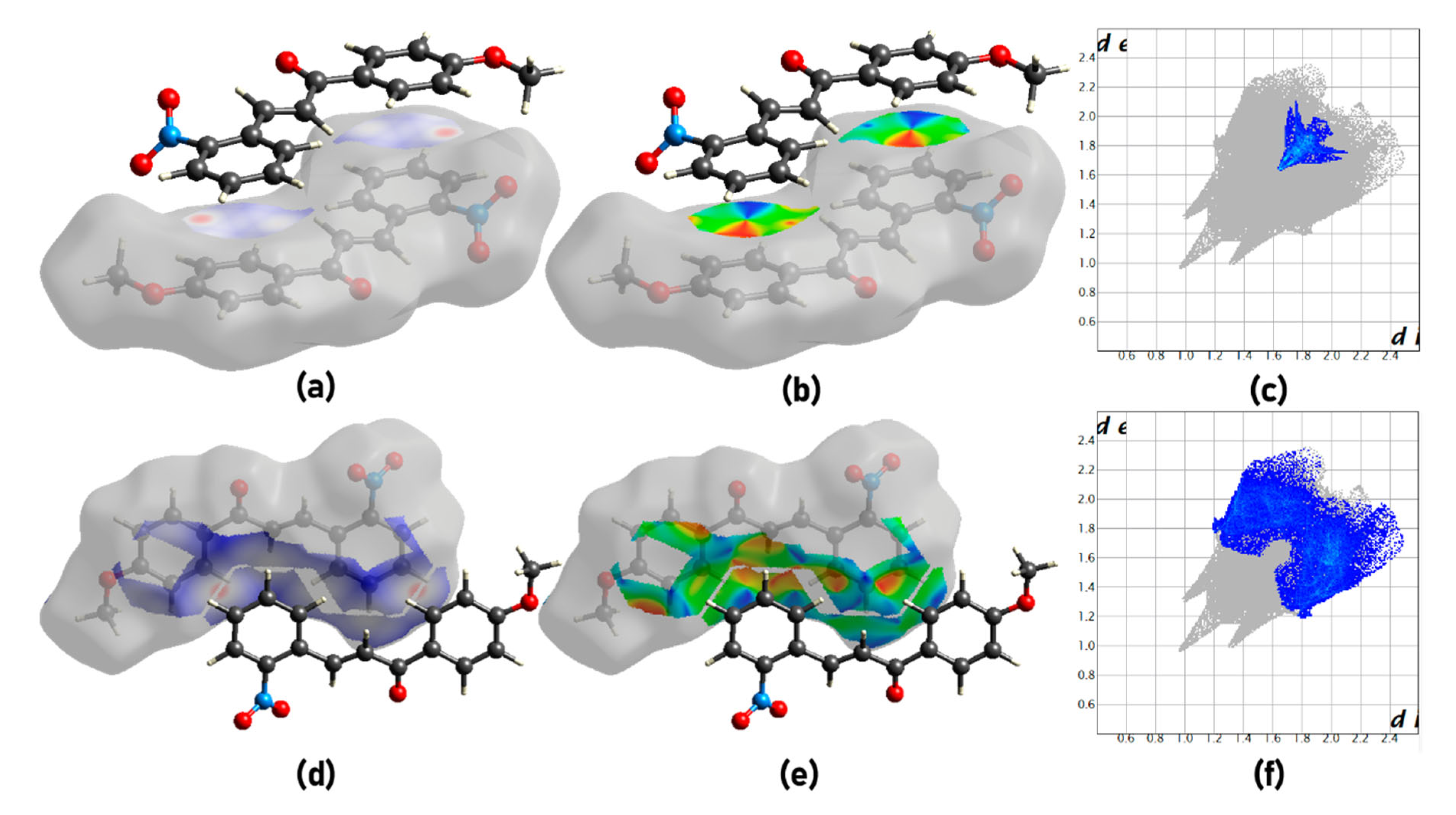
2.3. Hirshfeld Surface Analysis
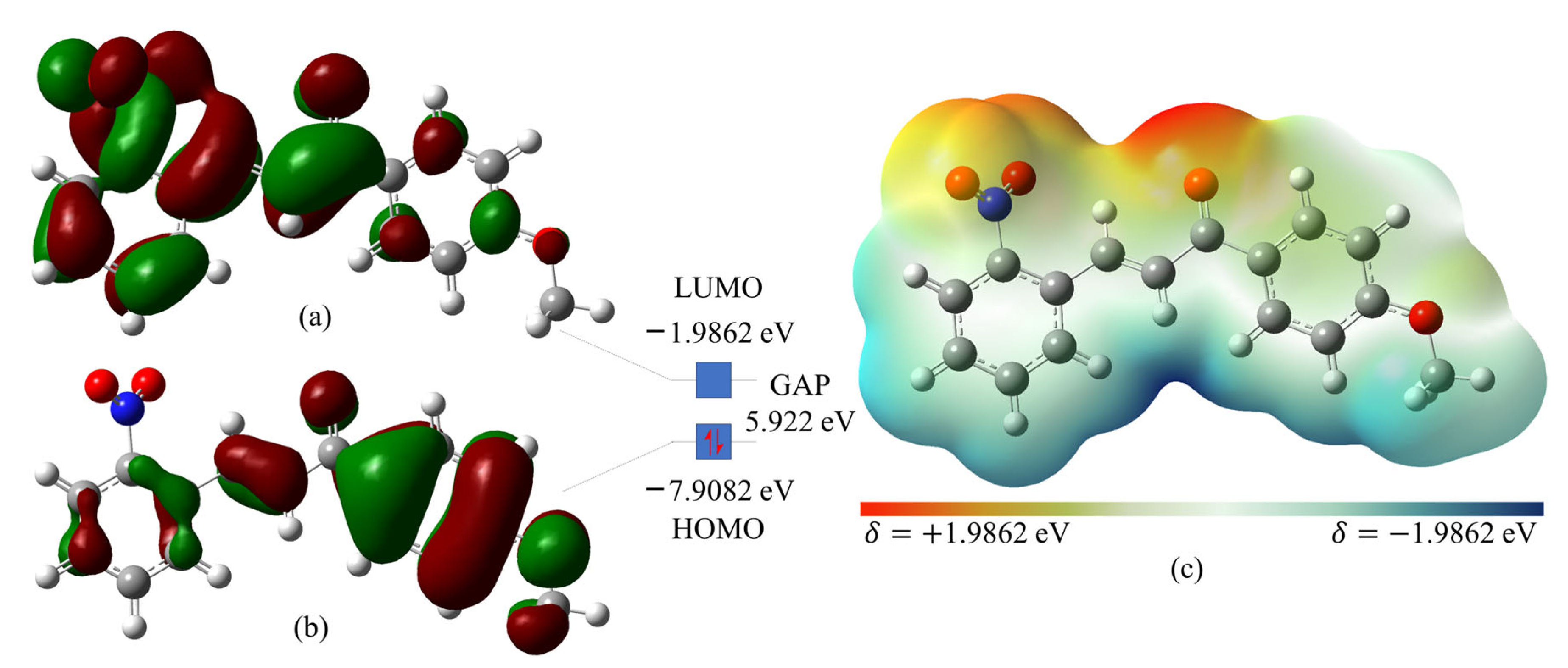
2.4. Theoretical Calculations
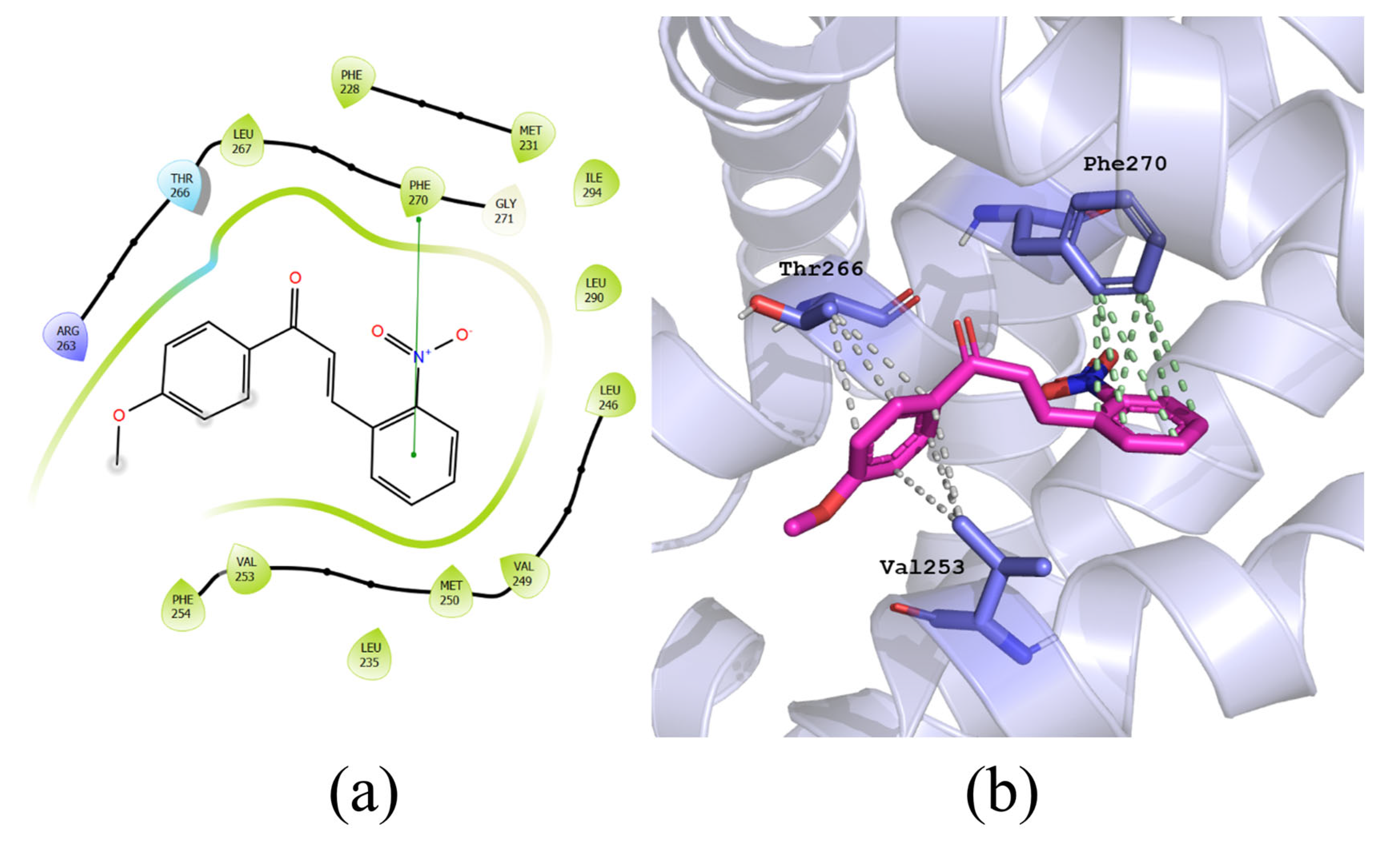
2.5. Molecular Docking
2.6. Experimental Assays
2.6.1. Evaluation of Antiproliferative Activity In Vitro Using MTT Method
2.6.2. Evaluation of Antiproliferative Activity Using the Trypan Blue Exclusion Method
2.6.3. Morphological Analysis Using Differential Staining with Rapid Panoptic
3. Results and Discussion
3.1. Structural Analysis
3.2. Theoretical Calculations
3.3. Molecular Docking
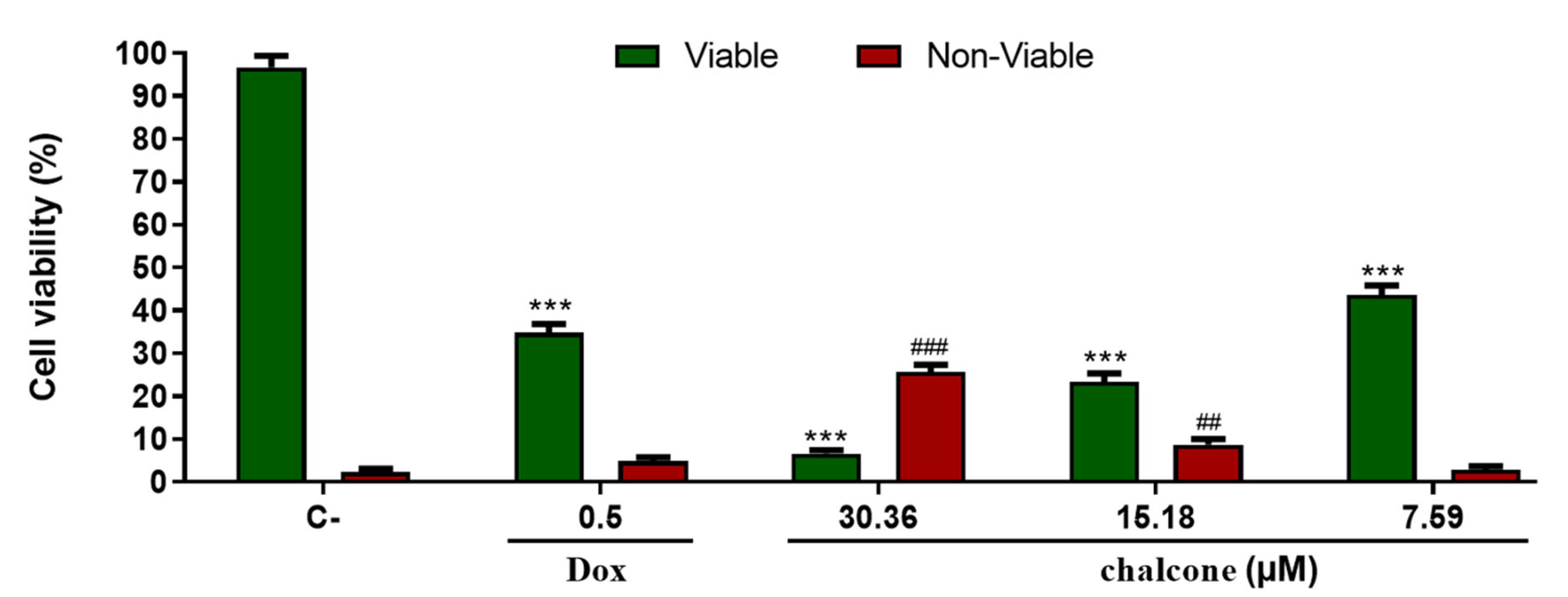
3.4. Cytotoxic Assay
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- McGuire, S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press. Adv. Nutr. 2016, 7, 418–419. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Report on Cancer: Setting Priorities, Investing Wisely and Providing Care for All; WHO Press: Geneva, Switzerland, 2020; ISBN 9789240001299. [Google Scholar]
- Huang, C.Y.; Ju, D.T.; Chang, C.F.; Muralidhar Reddy, P.; Velmurugan, B.K. A review on the effects of current chemotherapy drugs and natural agents in treating non-small cell lung cancer. Biomedicine 2017, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Delaney, G.; Jacob, S.; Featherstone, C.; Barton, M. The role of radiotherapy in cancer treatment: Estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer 2005, 104, 1129–1137. [Google Scholar] [CrossRef]
- Vaupel, P.; Thews, O.; Hoeckel, M. Treatment resistance of solid tumors: Role of hypoxia and anemia. Med. Oncol. 2001, 18, 243–259. [Google Scholar] [CrossRef]
- Lloyd, K.L.; Cree, I.A.; Savage, R.S. Prediction of resistance to chemotherapy in ovarian cancer: A systematic review. BMC Cancer 2015, 15, 117. [Google Scholar] [CrossRef]
- Chen, Y.; Gibson, S.B. Mcl-1 is a Gate Keeper Regulating Cell Death in Cancer Cells. J. Clin. Exp. Oncol. 2017, 6. [Google Scholar] [CrossRef]
- Xiang, W.; Yang, C.Y.; Bai, L. MCL-1 inhibition in cancer treatment. Onco. Targets Ther. 2018, 11, 7301–7314. [Google Scholar] [CrossRef]
- Tong, J.; Wang, P.; Tan, S.; Chen, D.; Nikolovska-Coleska, Z.; Zou, F.; Yu, J.; Zhang, L. Mcl-1 Degradation Is Required for Targeted Therapeutics to Eradicate Colon Cancer Cells. Cancer Res. 2017, 77, 2512–2521. [Google Scholar] [CrossRef]
- Akgul, C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell. Mol. Life Sci. 2009, 66, 1326–1336. [Google Scholar] [CrossRef]
- Campbell, K.J.; Dhayade, S.; Ferrari, N.; Sims, A.H.; Johnson, E.; Mason, S.M.; Dickson, A.; Ryan, K.M.; Kalna, G.; Edwards, J.; et al. MCL-1 is a prognostic indicator and drug target in breast cancer article. Cell Death Dis. 2018, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Belmar, J.; Fesik, S.W. Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacol. Ther. 2015, 145, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Leverson, J.D.; Zhang, H.; Chen, J.; Tahir, S.K.; Phillips, D.C.; Xue, J.; Nimmer, P.; Jin, S.; Smith, M.; Xiao, Y.; et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Dis. 2015, 6, e1590. [Google Scholar] [CrossRef]
- Young, A.I.J.; Law, A.M.K.; Castillo, L.; Chong, S.; Cullen, H.D.; Koehler, M.; Herzog, S.; Brummer, T.; Lee, E.F.; Fairlie, W.D.; et al. MCL-1 inhibition provides a new way to suppress breast cancer metastasis and increase sensitivity to dasatinib. Breast Cancer Res. 2016, 18, 125. [Google Scholar] [CrossRef] [PubMed]
- Hird, A.W.; Tron, A.E. Recent advances in the development of Mcl-1 inhibitors for cancer therapy. Pharmacol. Ther. 2019, 198, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Magis, A.T.; Xu, K.; Park, D.; Yu, D.S.; Owonikoko, T.K.; Sica, G.L.; Satola, S.W.; Ramalingam, S.S.; Curran, W.J.; et al. Targeting Mcl-1 enhances DNA replication stress sensitivity to cancer therapy. J. Clin. Investig. 2018, 128, 500–516. [Google Scholar] [CrossRef]
- Yamaguchi, R.; Lartigue, L.; Perkins, G. Targeting Mcl-1 and other Bcl-2 family member proteins in cancer therapy. Pharmacol. Ther. 2019, 195, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhou, D.; Li, X.Z.; Jiang, Z.; Tan, C.; Wei, X.Y.; Ling, J.; Jing, J.; Liu, F.; Li, N. A natural chalcone induces apoptosis in lung cancer cells: 3D-QSAR, docking and an in vivo/vitro assay. Sci. Rep. 2017, 7, 10729. [Google Scholar] [CrossRef]
- Rivera, A.G.; Mariscal, H.A.; Ceronio, N.R.; Fuente, L.F.R.; García, C.E.L. Synthesis and anti-inflammatory activity of three nitro chalcones. Bioorg. Med. Chem. Lett. 2013, 23, 5519–5522. [Google Scholar] [CrossRef]
- Tang, L.; Sun, L.; Zhang, Y.; Hu, L. Methoxy Chalcones: An Overview of Their Chemical Structure, Antioxidant Activity, and Mechanisms Involved in Neuroprotection. Curr. Pharm. Des. 2019, 25, 2084–2092. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Bruno, I.J.; Cole, J.C.; Edgington, P.R.; Kessler, M.; Macrae, C.F.; McCabe, P.; Pearson, J.; Taylor, R. New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 389–397. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Farrugia, L. ORTEP-3 for Windows—A version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef]
- Bruno, I.J.; Cole, J.C.; Kessler, M.; Luo, J.; Motherwell, W.D.S.; Purkis, L.H.; Smith, B.R.; Taylor, R.; Cooper, R.I.; Harris, S.E.; et al. Retrieval of Crystallographically-Derived Molecular Geometry Information. J. Chem. Inf. Comput. Sci. 2004, 44, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Groom, C.R.; Allen, F.H. The Cambridge Structural Database in retrospect and prospect. Angew. Chemie Int. Ed. 2014, 53, 662–671. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, J.J.; Spackman, M.A.; Mitchell, A.S. Novel tools for visualizing and exploring intermolecular interactions in molecular crystals. Acta Crystallogr. Sect. B Struct. Sci. 2004, 60, 627–668. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 7, 3814–3816. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Turner, M.J.; Jayatilaka, D.; Spackman, M.A.; Grimwood, D.J.; Wolff, S.K. Crystal Explorer, Version 3.1; University of Western Australia: Perth, Australia, 2012.
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
- Tirado-Rives, J.; Jorgensen, W.L. Performance of B3LYP density functional methods for a large set of organic molecules. J. Chem. Theory Comput. 2008, 4, 297–306. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Dennington, R.; Keith, T.; Millam, J. GaussView, Version 5; Semichem Inc.: Shawnee, KS, USA, 2009.
- MacRae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Lee, T.; Christov, P.P.; Shaw, S.; Tarr, J.C.; Zhao, B.; Veerasamy, N.; Jeon, K.O.; Mills, J.J.; Bian, Z.; Sensintaffar, J.L.; et al. Discovery of Potent Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors That Demonstrate in Vivo Activity in Mouse Xenograft Models of Human Cancer. J. Med. Chem. 2019, 62, 3971–3988. [Google Scholar] [CrossRef]
- Schrodinger. Maestro; Version 11.2; Schrödinger LLC.: New York, NY, USA, 2016. [Google Scholar]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- OpenEye. PICTO; Version 4.4.0.4; OpenEye Scientific Software: Santa Fe, NM, USA, 2018. [Google Scholar]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Nowakowska, Z. A review of anti-infective and anti-inflammatory chalcones. Eur. J. Med. Chem. 2007, 42, 125–137. [Google Scholar] [CrossRef]
- Menezes, A.P.; Jayarama, A.; Ng, S.W. Crucial role of molecular planarity on the second order nonlinear optical property of pyridine based chalcone single crystals. J. Mol. Struct. 2015, 1088, 85–94. [Google Scholar] [CrossRef]
- Perepichka, D.F.; Bryce, M.R. Molecules with exceptionally small HOMO-LUMO gaps. Angew. Chemie Int. Ed. 2005, 44, 5370–5373. [Google Scholar] [CrossRef] [PubMed]
- Custodio, J.M.F.; Vaz, W.F.; de Andrade, F.M.; Camargo, A.J.; Oliveira, G.R.; Napolitano, H.B. Substitution effect on a hydroxylated chalcone: Conformational, topological and theoretical studies. J. Mol. Struct. 2017, 1136, 69–79. [Google Scholar] [CrossRef]
- Carvalho, P.S.; Custodio, J.M.F.; Vaz, W.F.; Cirilo, C.C.; Cidade, A.F.; Aquino, G.L.B.; Campos, D.M.B.; Cravo, P.; Coelho, C.J.; Oliveira, S.S.; et al. Conformation analysis of a novel fluorinated chalcone. J. Mol. Model. 2017, 23, 97. [Google Scholar] [CrossRef] [PubMed]
- Denis, C.; Sopková-De Oliveira Santos, J.; Bureau, R.; Voisin-Chiret, A.S. Hot-Spots of Mcl-1 Protein. J. Med. Chem. 2020, 63, 928–943. [Google Scholar] [CrossRef]
- Huang, N.; Shoichet, B.K.; Irwin, J.J. Benchmarking Sets for Molecular Docking. J. Med. Chem. 2006, 49, 6789–6801. [Google Scholar] [CrossRef] [PubMed]
- Bleicher, K.H.; Böhm, H.-J.; Müller, K.; Alanine, A.I. Hit and lead generation: Beyond high-throughput screening. Nat. Rev. Drug Discov. 2003, 2, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Raha, K.; Merz, K.M.; Roux, B. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des. 2007, 21, 281–306. [Google Scholar] [CrossRef]






| Chemical formula | C16H13NO4 |
| Mr | 283.27 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 120 |
| a, b, c (Å) | 12.4534 (19), 6.8905 (10), 15.555 (2) |
| β (°) | 101.834 (2) |
| V (Å3) | 1306.4 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.11 |
| Crystal size (mm) | 0.32 × 0.23 × 0.22 |
| No. of measured, independent, and observed [I > 2σ(I)] reflections | 23943, 3233, 2703 |
| Rint | 0.032 |
| (sin θ/λ)max (Å−1) | 0.666 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.037, 0.105, 1.04 |
| No. of reflections | 3233 |
| No. of parameters | 194 |
| H-atom treatment | H atoms treated with a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.34, −0.25 |
| Dihedral Angle | Atoms | Value (°) |
|---|---|---|
| O2—N1—C1—C6 | 44.0 (2) | |
| C1—C6—C7—C8 | 161.7 (1) | |
| C6—C7—C8—C9 | 174.8 (1) | |
| C7—C8—C9—C10 | 168.5 (1) | |
| C8—C9—C10—C15 | 167.9 (1) | |
| O3—C9—O4—C16 | 173.8 (1) | |
| C14—C13—O4—C16 | 172.7 (1) |
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| C7—H7···O2 i | 0.97 (1) | 2.28 (1) | 2.8929 (15) | 121 (1) |
| C2—H2···O3 ii | 0.95 | 2.50 | 3.3750 (15) | 152 |
| C3—H3···O4 iii | 0.95 | 2.47 | 3.4179 (15) | 177 |
| C5—H5···O1 iv | 0.95 | 2.64 | 3.324 (15) | 129 |
| C11—H11···O2 iv | 0.95 | 2.42 | 3.2092 (15) | 141 |
| Glide Score | RMSD | |
|---|---|---|
| 4′-methoxy-2-nitrochalcone | −8.571 | - |
| Redocking/reference ligand | −15.212 | 1.1 |
| IC50 (µM)—24 h | IC50 (µM)—48 h | IC50 (µM)—72 h |
| 15.18 (14.21–16.23) | 8.69 (8.19–9.23) | 7.79 (7.54–8.05) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, I.V.; Naves, L.F.N.; Custodio, J.M.F.; Vidal, H.D.A.; Queiroz, J.E.; Oliver, A.G.; Borba, J.V.B.; Neves, B.J.; Brito, L.M.; Pessoa, C.; et al. A Structure-Guided Designed Small Molecule Is an Anticancer Agent and Inhibits the Apoptosis-Related MCL-1 Protein. Biophysica 2023, 3, 446-462. https://doi.org/10.3390/biophysica3030029
Machado IV, Naves LFN, Custodio JMF, Vidal HDA, Queiroz JE, Oliver AG, Borba JVB, Neves BJ, Brito LM, Pessoa C, et al. A Structure-Guided Designed Small Molecule Is an Anticancer Agent and Inhibits the Apoptosis-Related MCL-1 Protein. Biophysica. 2023; 3(3):446-462. https://doi.org/10.3390/biophysica3030029
Chicago/Turabian StyleMachado, Ingrid V., Luiz F. N. Naves, Jean M. F. Custodio, Hérika D. A. Vidal, Jaqueline E. Queiroz, Allen G. Oliver, Joyce V. B. Borba, Bruno J. Neves, Lucas M. Brito, Claudia Pessoa, and et al. 2023. "A Structure-Guided Designed Small Molecule Is an Anticancer Agent and Inhibits the Apoptosis-Related MCL-1 Protein" Biophysica 3, no. 3: 446-462. https://doi.org/10.3390/biophysica3030029
APA StyleMachado, I. V., Naves, L. F. N., Custodio, J. M. F., Vidal, H. D. A., Queiroz, J. E., Oliver, A. G., Borba, J. V. B., Neves, B. J., Brito, L. M., Pessoa, C., Napolitano, H. B., & Aquino, G. L. B. d. (2023). A Structure-Guided Designed Small Molecule Is an Anticancer Agent and Inhibits the Apoptosis-Related MCL-1 Protein. Biophysica, 3(3), 446-462. https://doi.org/10.3390/biophysica3030029








