A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis, and Primary Central Nervous System Lymphoma: An External Validation Study
Abstract
1. Introduction
2. Methods
2.1. Study Definition
2.2. Patient Selection
2.3. MR Acquisition and Image Pre-Processing
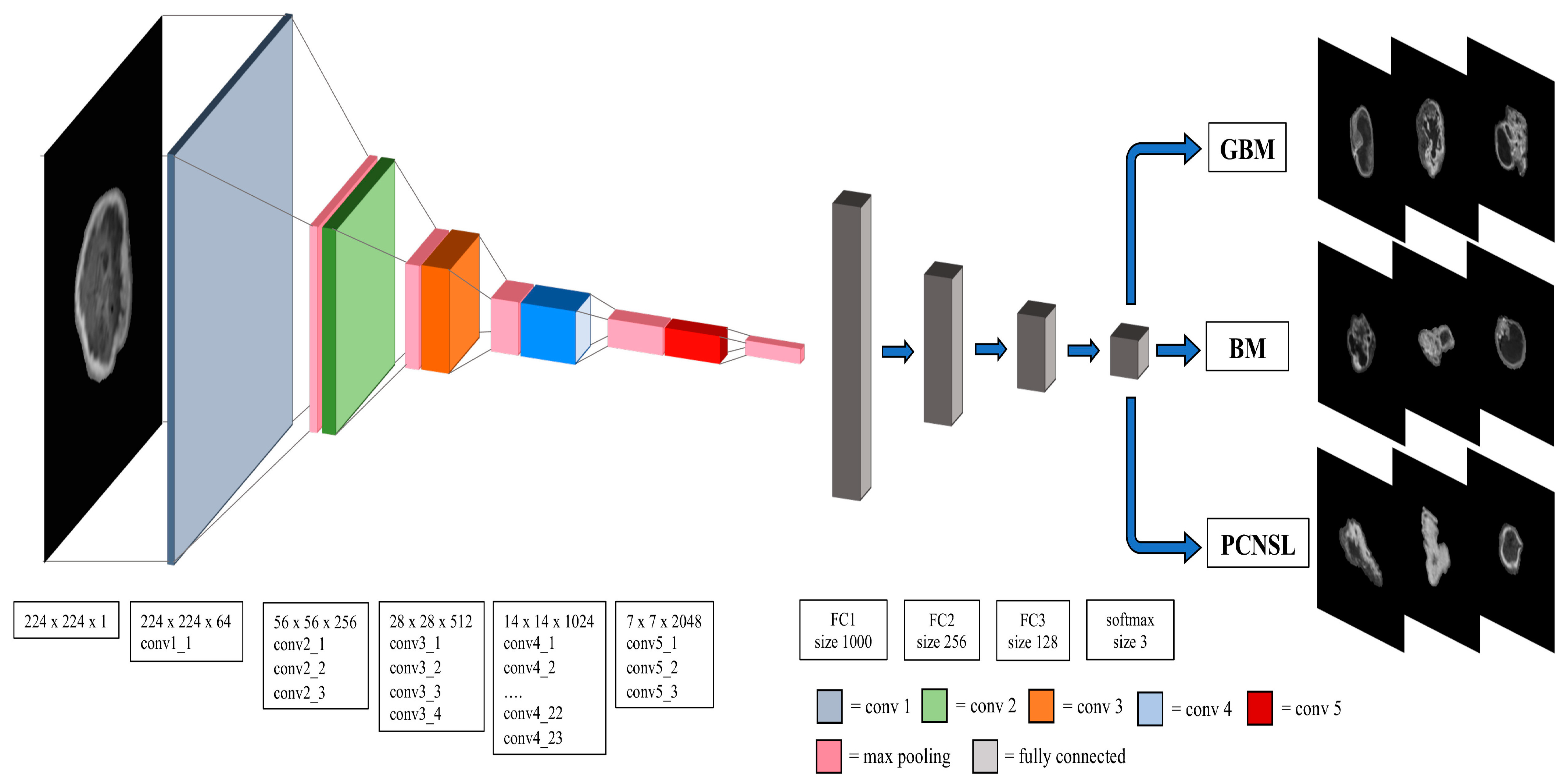
2.4. Convolutional Neural Network Model
2.5. Performance Metrics
2.6. Human “Gold Standard” Performance
2.7. Software and Hardware
3. Results
3.1. DNN Model Performance Metrics Evaluation
3.2. Comparison of DNN Model and Neuroradiologists’ Gold Standard Performance
4. Discussion
4.1. Performance Validation
4.2. Perspective for Clinical Application and Public Health Impact
4.3. Perspective in Medical Education
4.4. Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-Oncol. 2018, 20, iv1–iv86. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro-Oncol. 2020, 22, iv1–iv96. [Google Scholar] [CrossRef]
- Nayak, L.; Lee, E.Q.; Wen, P.Y. Epidemiology of brain metastases. Curr. Oncol. Rep. 2012, 14, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A Survey of Brain Tumor Segmentation and Classification Algorithms. J. Imaging 2021, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Abd-Ellah, M.K.; Awad, A.I.; Khalaf, A.A.M.; Hamed, H.F.A. A review on brain tumor diagnosis from MRI images: Practical implications, key achievements, and lessons learned. Magn. Reson. Imaging 2019, 61, 300–318. [Google Scholar] [CrossRef]
- Baris, M.M.; Celik, A.O.; Gezer, N.S.; Ada, E. Role of mass effect, tumor volume and peritumoral edema volume in the differential diagnosis of primary brain tumor and metastasis. Clin. Neurol. Neurosurg. 2016, 148, 67–71. [Google Scholar] [CrossRef]
- Batchelor, T.; Loeffler, J.S. Primary CNS lymphoma. J. Clin. Oncol. 2006, 24, 1281–1288. [Google Scholar] [CrossRef]
- Augustin Toma, M.; Gerhard-Paul Diller, M.P.; Patrick, R.; Lawler, M.M. Deep Learning in Medicine. JACC Adv. 2022, 1, 100017. [Google Scholar] [CrossRef]
- Kim, M.; Yun, J.; Cho, Y.; Shin, K.; Jang, R.; Bae, H.J.; Kim, N. Deep Learning in Medical Imaging. Neurospine 2019, 16, 657–668. [Google Scholar] [CrossRef]
- Lee, W.-J.; Hong, S.D.; Woo, K.I.; Seol, H.J.; Choi, J.W.; Lee, J.-I.; Nam, D.-H.; Kong, D.-S. Combined endoscopic endonasal and transorbital multiportal approach for complex skull base lesions involving multiple compartments. Acta Neurochir. 2022, 164, 1911–1922. [Google Scholar] [CrossRef]
- Zaharchuk, G.; Gong, E.; Wintermark, M.; Rubin, D.; Langlotz, C.P. Deep Learning in Neuroradiology. AJNR. Am. J. Neuroradiol. 2018, 39, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Tariciotti, L.; Palmisciano, P.; Giordano, M.; Remoli, G.; Lacorte, E.; Bertani, G.; Locatelli, M.; Dimeco, F.; Caccavella, V.M.; Prada, F. Artificial intelligence-enhanced intraoperative neurosurgical workflow: State of the art and future perspectives. J. Neurosurg. Sci. 2021, 66, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Tariciotti, L.; Caccavella, V.M.; Fiore, G.; Schisano, L.; Carrabba, G.; Borsa, S.; Giordano, M.; Palmisciano, P.; Remoli, G.; Remore, L.G.; et al. A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis and Primary Central Nervous System Lymphoma: A Pilot Study. Front. Oncol. 2022, 12, 816638. [Google Scholar] [CrossRef] [PubMed]
- Simera, I.; Moher, D.; Hoey, J.; Schulz, K.F.; Altman, D.G. The EQUATOR Network and reporting guidelines: Helping to achieve high standards in reporting health research studies. Maturitas 2009, 63, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Sounderajah, V.; Ashrafian, H.; Aggarwal, R.; De Fauw, J.; Denniston, A.K.; Greaves, F.; Karthikesalingam, A.; King, D.; Liu, X.; Markar, S.R.; et al. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: The STARD-AI Steering Group. Nat. Med. 2020, 26, 807–808. [Google Scholar] [CrossRef]
- Cohen, J.F.; Korevaar, D.A.; Altman, D.G.; Bruns, D.E.; Gatsonis, C.A.; Hooft, L.; Irwig, L.; Levine, D.; Reitsma, J.B.; De Vet, H.C.W.; et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: Explanation and elaboration. BMJ Open 2016, 6, e012799. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. IEEE Conf. Comput. Vis. Pattern Recognit. 2009, 248–255. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Larroza, A.; Bodí, V.; Moratal, D. Texture Analysis in Magnetic Resonance Imaging: Review and Considerations for Future Applications. In Assessment of Cellular and Organ Function and Dysfunction Using Direct and Derived MRI Methodologies; IntechOpen Limited: London, UK, 2016. [Google Scholar] [CrossRef]
- Kunimatsu, A.; Kunimatsu, N.; Yasaka, K.; Akai, H.; Kamiya, K.; Watadani, T.; Mori, H.; Abe, O. Machine learning-based texture analysis of contrast-enhanced mr imaging to differentiate between glioblastoma and primary central nervous system lymphoma. Magn. Reson. Med. Sci. 2019, 18, 44–52. [Google Scholar] [CrossRef]
- Fruehwald-Pallamar, J.; Hesselink, J.; Mafee, M.; Holzer-Fruehwald, L.; Czerny, C.; Mayerhoefer, M. Texture-Based Analysis of 100 MR Examinations of Head and Neck Tumors—Is It Possible to Discriminate Between Benign and Malignant Masses in a Multicenter Trial? In RöFo-Fortschritte auf dem Gebiet der Röntgenstrahlen und der Bildgeb. Verfahren; Thieme: New York, NY, USA, 2015; Volume 188, pp. 195–202. [Google Scholar] [CrossRef]
- Tiwari, P.; Prasanna, P.; Rogers, L.; Wolansky, L.; Badve, C.; Sloan, A.; Cohen, M.; Madabhushi, A. Texture descriptors to distinguish radiation necrosis from recurrent brain tumors on multi-parametric MRI. In Proceedings of the Medical Imaging 2014: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2014; Volume 9035, p. 90352B. [Google Scholar]
- Xiao, D.-D.; Yan, P.-F.; Wang, Y.-X.; Osman, M.S.; Zhao, H.-Y. Glioblastoma and primary central nervous system lymphoma: Preoperative differentiation by using MRI-based 3D texture analysis. Clin. Neurol. Neurosurg. 2018, 173, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Davnall, F.; Yip, C.S.P.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.J.; Goh, V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Imaging 2012, 3, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.; Lupo, J.M.; Chen, M.H.; Lamborn, K.R.; McDermott, M.W.; Berger, M.S.; Nelson, S.J.; Dillon, W.P. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Am. J. Neuroradiol. 2007, 28, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Liang, D.; Zhang, Y.; Yao, W. Histogram analysis of absolute cerebral blood volume map can distinguish glioblastoma from solitary brain metastasis. Medicine 2019, 98, e17515. [Google Scholar] [CrossRef]
- Raza, S.M.; Lang, F.F.; Aggarwal, B.B.; Fuller, G.N.; Wildrick, D.M.; Sawaya, R. Necrosis and Glioblastoma: A Friend or a Foe? A Review and a Hypothesis. Neurosurgery 2002, 51, 2–13. [Google Scholar] [CrossRef]
- Thammaroj, J.; Wongwichit, N.; Boonrod, A. Evaluation of Perienhancing Area in Differentiation between Glioblastoma and Solitary Brain Metastasis. Asian Pac. J. Cancer Prev. 2020, 21, 2525. [Google Scholar] [CrossRef]
- Cossy-Gantner, A.; Germann, S.; Schwalbe, N.R.; Wahl, B. Artificial intelligence (AI) and global health: How can AI contribute to health in resource-poor settings? BMJ Glob. Health 2018, 3, 798. [Google Scholar] [CrossRef]
- Guo, J.; Li, B. The Application of Medical Artificial Intelligence Technology in Rural Areas of Developing Countries. Health Equity 2018, 2, 174. [Google Scholar] [CrossRef]
- Hoodbhoy, Z.; Hasan, B.; Siddiqui, K. Does artificial intelligence have any role in healthcare in low resource settings? J. Med. Artif. Intell. 2019, 2, 854. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Ben Hadj Hassen, A.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Kooi, T.; Litjens, G.; van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Kalmet, P.H.S.; Sanduleanu, S.; Primakov, S.; Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Hulst, L.; Lambin, P.; Poeze, M. Deep learning in fracture detection: A narrative review. Acta Orthop. 2020, 91, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.M.; Quittes, L.; Costa, P.H.V.; Ramos, T.M.; Rodrigues, A.C.F.; Nicolato, A.; Malheiros, J.A.; Machado, C. Computer vision coaching microsurgical laboratory training: PRIME (Proficiency Index in Microsurgical Education) proof of concept. Neurosurg. Rev. 2021, 45, 1601–1606. [Google Scholar] [CrossRef]
- Smith, P.; Tang, L.; Balntas, V.; Young, K.; Athanasiadis, Y.; Sullivan, P.; Hussain, B.; Saleh, G.M. “PhacoTracking”: An evolving paradigm in ophthalmic surgical training. JAMA Ophthalmol. 2013, 131, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.Z.; Luengo, I.; Barbarisi, S.; Addis, C.; Culshaw, L.; Dorward, N.L.; Haikka, P.; Jain, A.; Kerr, K.; Koh, C.H.; et al. Automated operative workflow analysis of endoscopic pituitary surgery using machine learning: Development and preclinical evaluation (IDEAL stage 0). J. Neurosurg. 2021, 1–8. [Google Scholar] [CrossRef]
- Kitaguchi, D.; Takeshita, N.; Matsuzaki, H.; Oda, T.; Watanabe, M.; Mori, K.; Kobayashi, E.; Ito, M. Automated laparoscopic colorectal surgery workflow recognition using artificial intelligence: Experimental research. Int. J. Surg. 2020, 79, 88–94. [Google Scholar] [CrossRef]
- Ward, T.M.; Hashimoto, D.A.; Ban, Y.; Rattner, D.W.; Inoue, H.; Lillemoe, K.D.; Rus, D.L.; Rosman, G.; Meireles, O.R. Automated operative phase identification in peroral endoscopic myotomy. Surg. Endosc. 2021, 35, 4008–4015. [Google Scholar] [CrossRef]

| Glioblastoma | BM | PCNSL | p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| Count (N%) | Mean (SD) | Count (N%) | Mean (SD) | Count (N%) | Mean (SD) | |||
| Gender | Female | 26 (41.3%) | 12 (36.4%) | 8.0 (29.6%) | p > 0.05 | |||
| Male | 37 (58.7%) | 21 (63.6%) | 19.0 (70.4%) | p > 0.05 | ||||
| Age (years) | 64.4 (9.04) | 62.7 (14.2) | 58.5 (16.5) | p > 0.05 | ||||
| N° Slices of T1Gd sequence (N) | 108.0 (52.0) | 107.0 (59.0) | 74.0 (61.0) | p > 0.05 | ||||
| N° Slices of ROI (N) | 28.0 (19.0) | 21.0 (4.0) | 15.0 (14.0) | p > 0.05 | ||||
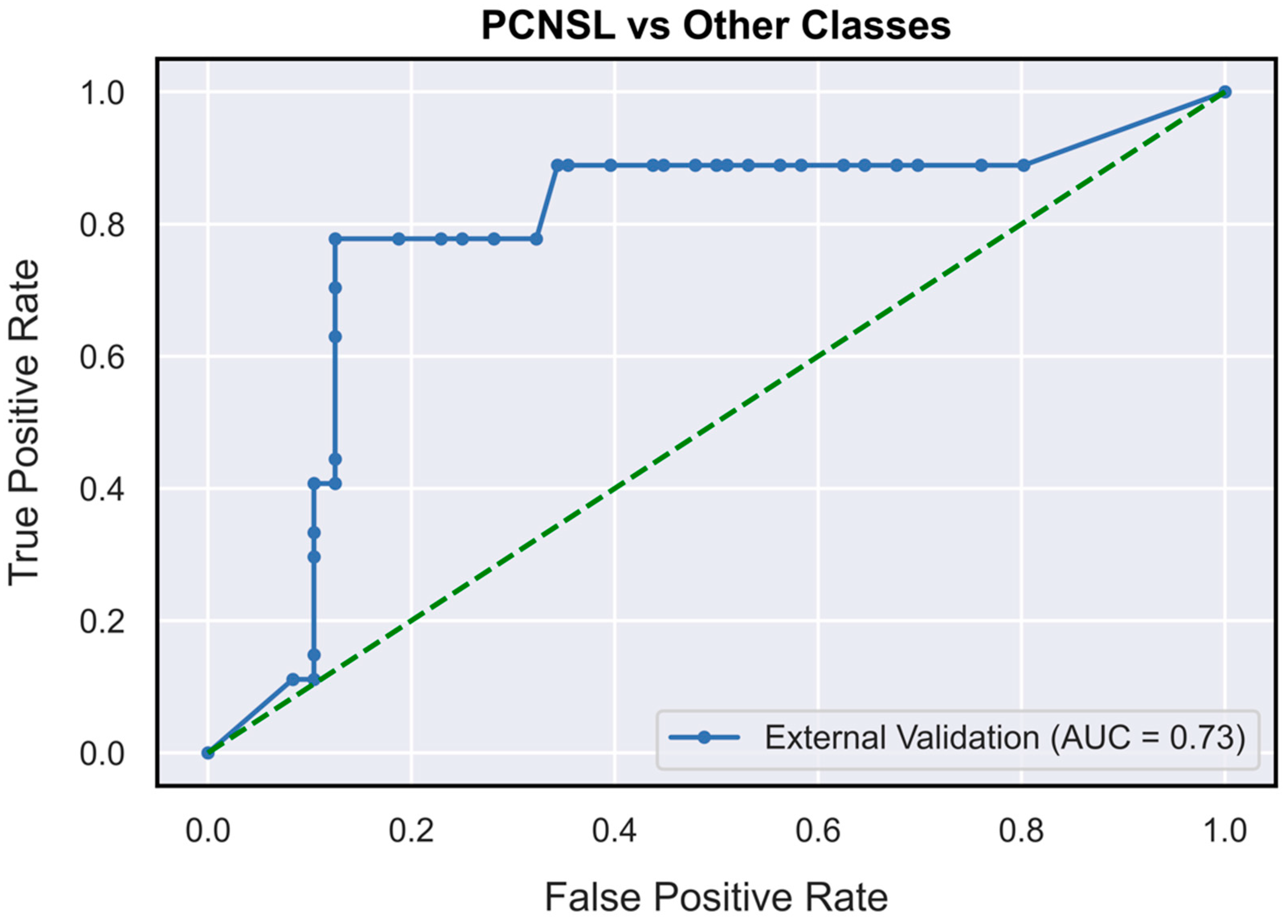
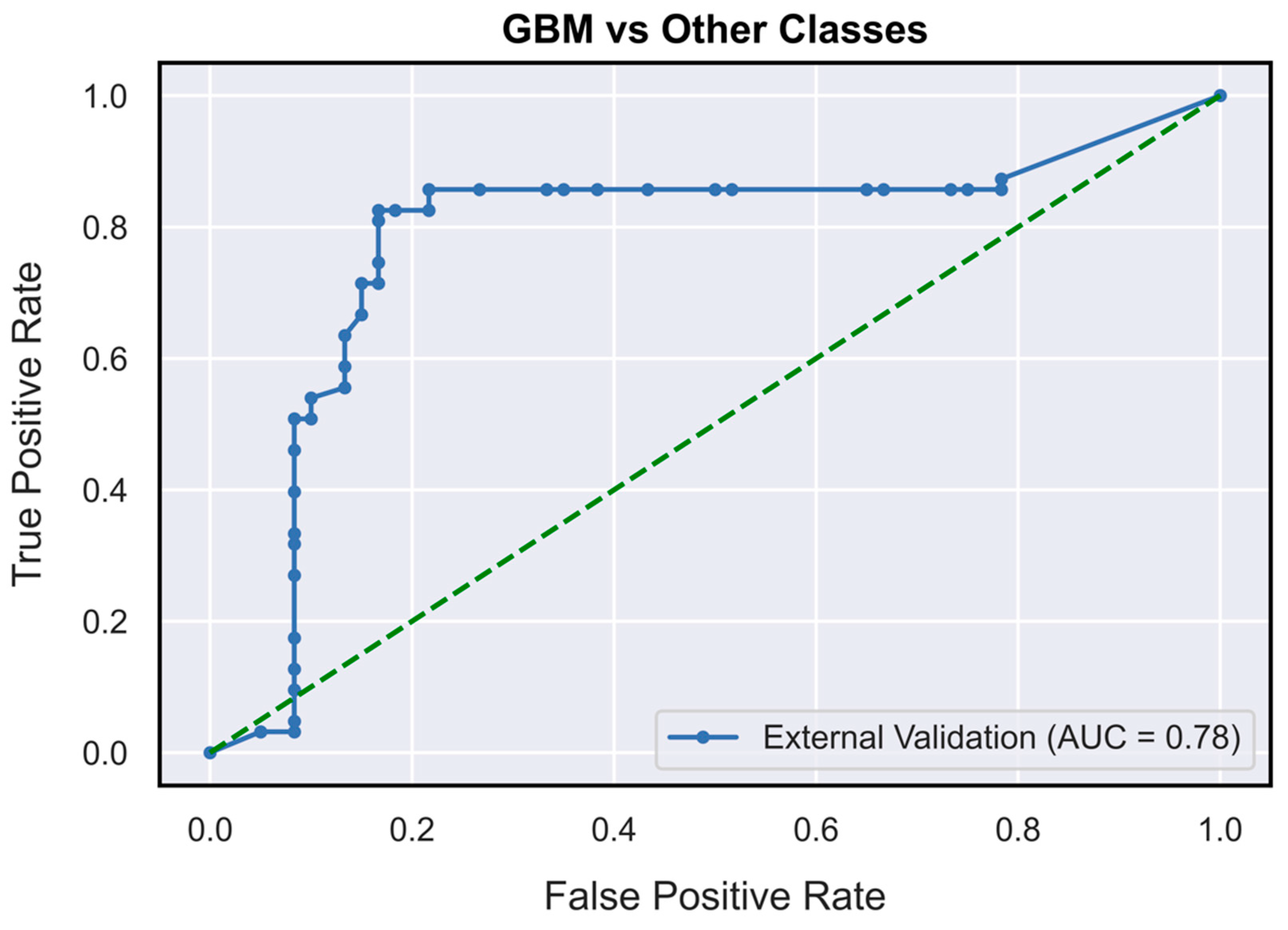
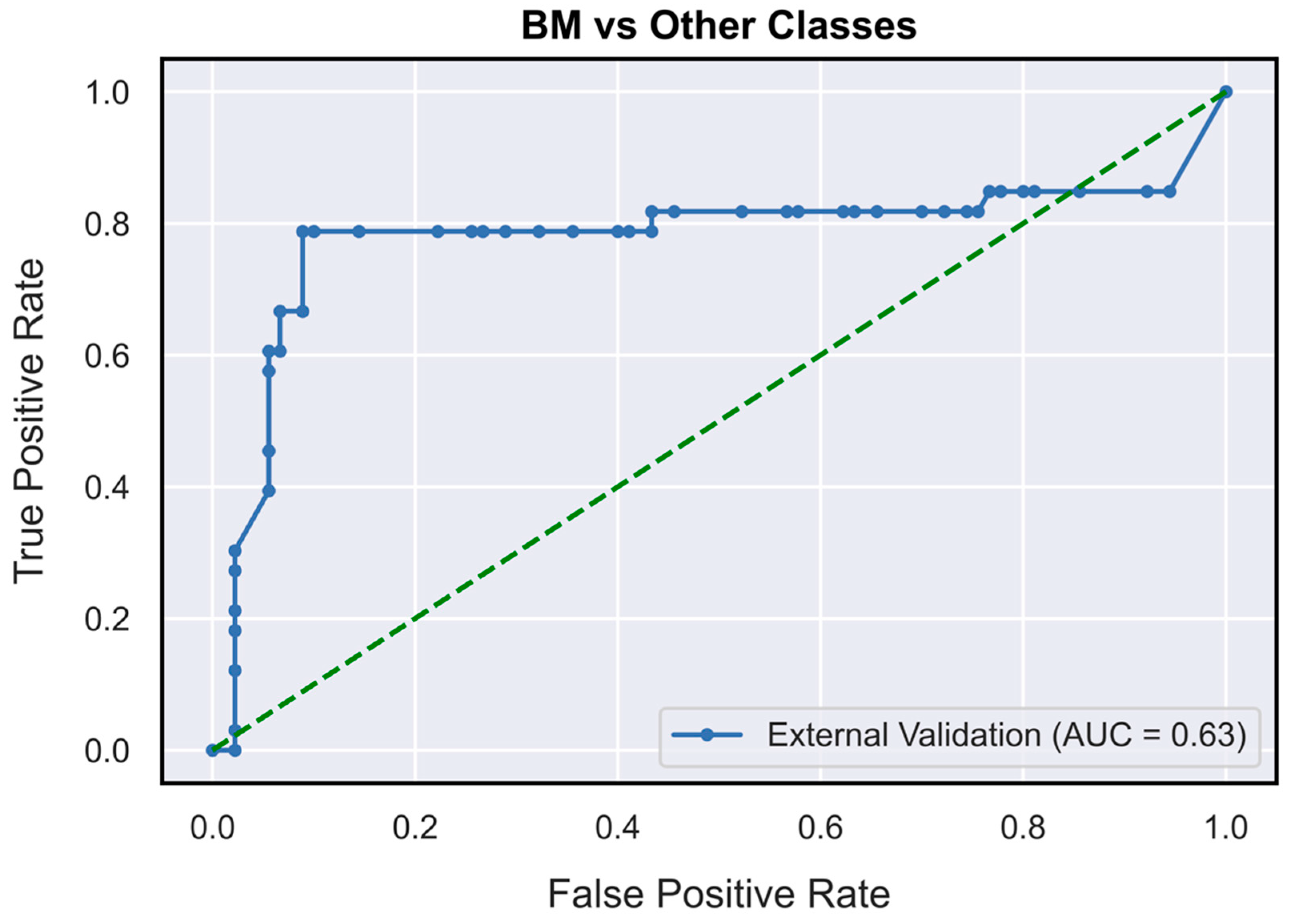
| Performance Metrics | PCNSL | Glioblastoma | BM |
|---|---|---|---|
| AUC | 0.73 (0.62–0.85) | 0.78 (0.71–0.87) | 0.63 (0.52–0.76) |
| Accuracy | 80.46% (74.8–87.01%) | 80.37% (74.8–86.99%) | 77.12% (71.54–83.74%) |
| Precision (PPV) | 54.85% (44.11–70.00%) | 84.13% (77.97–92.0%) | 57.71% (46.67–72.73%) |
| Recall (Sensitivity) | 66.86% (51.85–85.19%) | 76.14% (66.67–85.71%) | 57.04% (42.42–72.73%) |
| Specificity | 84.29% (78.12–91.67%) | 84.8% (78.33–93.33%) | 84.49% (77.78–91.14%) |
| F1-Score | 0.60 (0.50–0.73) | 0.80 (0.73–0.87) | 0.57 (0.45–0.70) |
| Performance Metrics | PCNSL | Glioblastoma | BM |
|---|---|---|---|
| Accuracy | 82.90% | 84,09% | 89.69% |
| Precision (PPV) | 65.21% | 87.50% | 79.31% |
| Negative predictive value (NPV) | 87.23% | 81.57% | 94.11% |
| Recall (Sensitivity) | 55.55% | 77.77% | 85.18% |
| Specificity | 91.11% | 89.85% | 91.42% |
| F1-Score | 0,595 | 0,819 | 0,818 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tariciotti, L.; Ferlito, D.; Caccavella, V.M.; Di Cristofori, A.; Fiore, G.; Remore, L.G.; Giordano, M.; Remoli, G.; Bertani, G.; Borsa, S.; et al. A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis, and Primary Central Nervous System Lymphoma: An External Validation Study. NeuroSci 2023, 4, 18-30. https://doi.org/10.3390/neurosci4010003
Tariciotti L, Ferlito D, Caccavella VM, Di Cristofori A, Fiore G, Remore LG, Giordano M, Remoli G, Bertani G, Borsa S, et al. A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis, and Primary Central Nervous System Lymphoma: An External Validation Study. NeuroSci. 2023; 4(1):18-30. https://doi.org/10.3390/neurosci4010003
Chicago/Turabian StyleTariciotti, Leonardo, Davide Ferlito, Valerio M. Caccavella, Andrea Di Cristofori, Giorgio Fiore, Luigi G. Remore, Martina Giordano, Giulia Remoli, Giulio Bertani, Stefano Borsa, and et al. 2023. "A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis, and Primary Central Nervous System Lymphoma: An External Validation Study" NeuroSci 4, no. 1: 18-30. https://doi.org/10.3390/neurosci4010003
APA StyleTariciotti, L., Ferlito, D., Caccavella, V. M., Di Cristofori, A., Fiore, G., Remore, L. G., Giordano, M., Remoli, G., Bertani, G., Borsa, S., Pluderi, M., Remida, P., Basso, G., Giussani, C., Locatelli, M., & Carrabba, G. (2023). A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis, and Primary Central Nervous System Lymphoma: An External Validation Study. NeuroSci, 4(1), 18-30. https://doi.org/10.3390/neurosci4010003








