Understanding Raman Spectral Based Classifications with Convolutional Neural Networks Using Practical Examples of Fungal Spores and Carotenoid-Pigmented Microorganisms
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Spores and Carotenoid-Containing Microorganisms
2.2. Sample Preparation
2.3. Spectral Recording
2.4. Data Preprocessing and Model Development
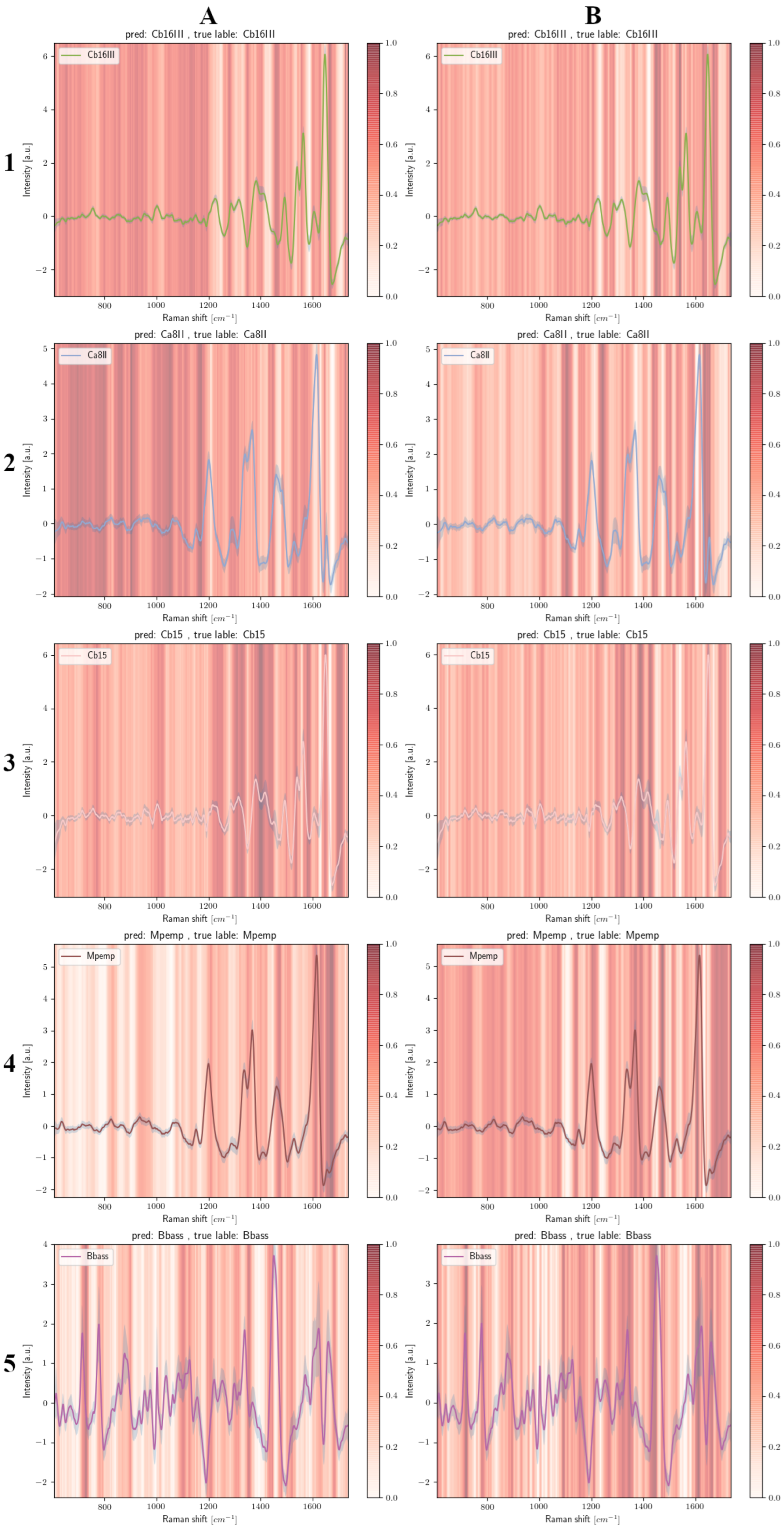
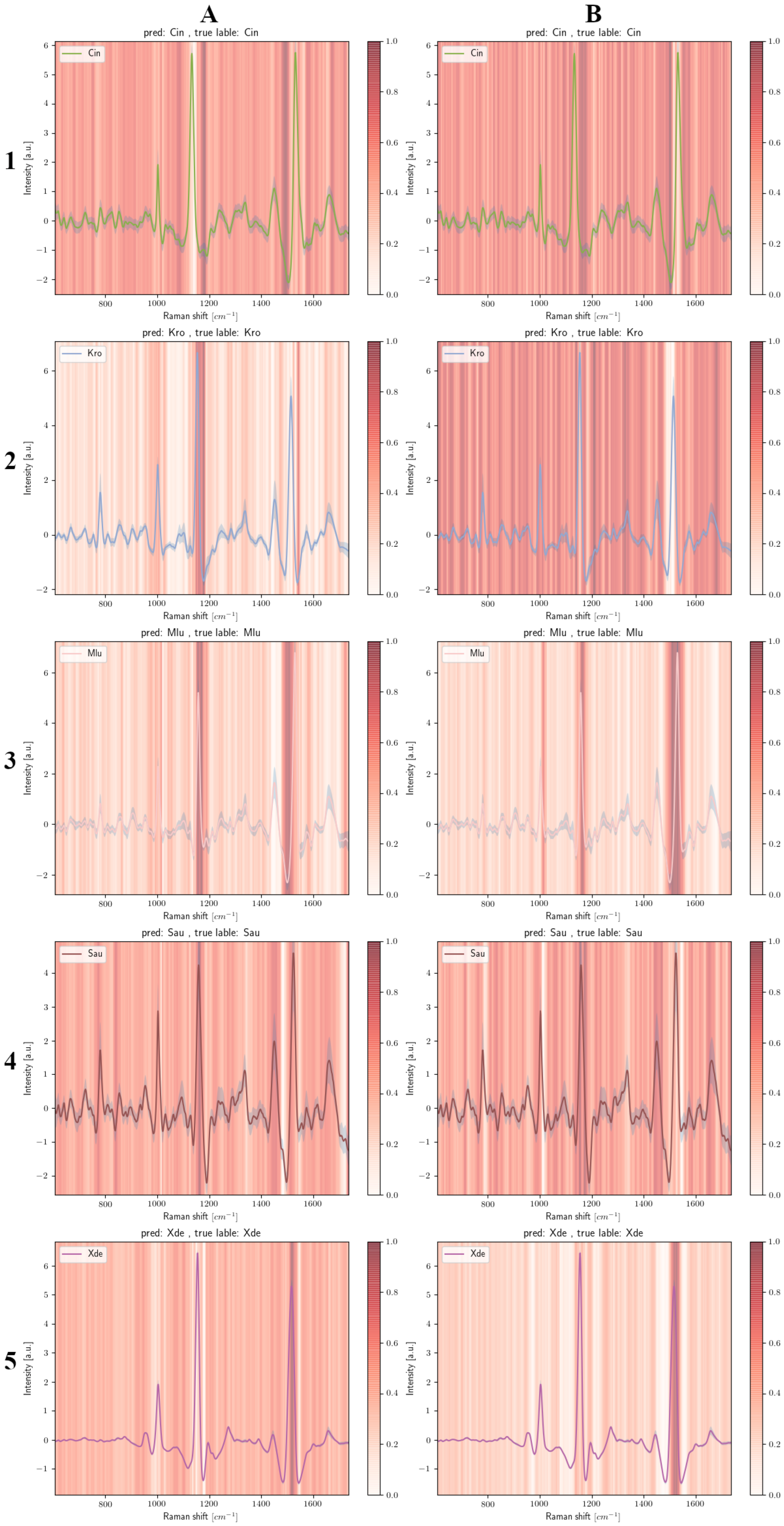
2.5. Grad-CAM
3. Results and Discussion
3.1. Raman Spectra Untreated and Preprocessed
3.2. PCA for General Estimation of the Classifiability
3.3. Predictive Models and Cross-Validation
3.4. Grad-CAM Results
3.4.1. Fungal Spores
3.4.2. Carotenoid-Containing Microorganisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stöckel, S.; Kirchhoff, J.; Neugebauer, U.; Rösch, P.; Popp, J. The application of Raman spectroscopy for the detection and identification of microorganisms. J. Raman Spectrosc. 2016, 47, 89–109. [Google Scholar] [CrossRef]
- Rösch, P.; Harz, M.; Krause, M.; Popp, J. Fast and reliable identification of microorganisms by means of Raman spectroscopy. In Biophotonics 2007: Optics in Life Science; Optical Society of America: Washington, DC, USA, 2007; pp. 6633–6645. [Google Scholar]
- Pahlow, S.; Meisel, S.; Cialla-May, D.; Weber, K.; Rösch, P.; Popp, J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv. Drug Deliv. Rev. 2015, 89, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef] [PubMed]
- Meisel, S.; Stöckel, S.; Elschner, M.; Melzer, F.; Rösch, P.; Popp, J. Raman spectroscopy as a potential tool for detection of Brucella spp. in milk. Appl. Environ. Microbiol. 2012, 78, 5575–5583. [Google Scholar] [CrossRef]
- Rösch, P.; Harz, M.; Schmitt, M.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Motzkus, H.W.; Lankers, M.; Hofer, S.; Thiele, H.; et al. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: Application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 2005, 71, 1626–1637. [Google Scholar] [CrossRef]
- Strola, S.A.; Baritaux, J.-C.; Schultz, E.; Simon, A.C.; Allier, C.; Espagnon, I.; Jary, D.; Dinten, J.M. Single bacteria identification by Raman spectroscopy. J. Biomed. Opt. 2014, 19, 111610. [Google Scholar] [CrossRef]
- Hetjens, B.T.; Tewes, T.J.; Platte, F.; Wichern, F. The application of Raman spectroscopy in identifying Metarhizium brunneum, Metarhizium pemphigi and Beauveria bassiana. Biocontrol Sci. Technol. 2021, 32, 329–340. [Google Scholar] [CrossRef]
- Tewes, T.J.; Kerst, M.; Platte, F.; Bockmühl, D.P. Raman Microscopic Identification of Microorganisms on Metal Surfaces via Support Vector Machines. Microorganisms 2022, 10, 556. [Google Scholar] [CrossRef]
- Kanno, N.; Kato, S.; Ohkuma, M.; Matsui, M.; Iwasaki, W.; Shigeto, S. Machine learning-assisted single-cell Raman fingerprinting for in situ and nondestructive classification of prokaryotes. iScience 2021, 24, 102975. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Popp, J. Vibrational spectroscopy-A powerful tool for the rapid identification of microbial cells at the single-cell level. Cytom. Part A 2009, 75, 104–113. [Google Scholar] [CrossRef]
- Mlynáriková, K.; Samek, O.; Bernatová, S.; Růžička, F.; Ježek, J.; Hároniková, A.; Šiler, M.; Zemánek, P.; Holá, V. Influence of culture media on microbial fingerprints using raman spectroscopy. Sensors 2015, 15, 29635–29647. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Popp, J. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst 2005, 130, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Hutsebaut, D.; Maquelin, K.; De Vos, P.; Vandenabeele, P.; Moens, L.; Puppels, G.J. Effect of Culture Conditions on the Achievable Taxonomic Resolution of Raman Spectroscopy Disclosed by Three Bacillus Species. Anal. Chem. 2004, 76, 6274–6281. [Google Scholar] [CrossRef] [PubMed]
- Bocklitz, T.; Walter, A.; Hartmann, K.; Rösch, P.; Popp, J. How to pre-process Raman spectra for reliable and stable models? Anal. Chim. Acta 2011, 704, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, W.; Stöckel, S.; Rösch, P.; Popp, J. Improving chemometric results by optimizing the dimension reduction for Raman spectral data sets. J. Raman Spectrosc. 2014, 45, 930–940. [Google Scholar] [CrossRef]
- Kumar, B.N.V.; Kampe, B.; Rösch, P.; Popp, J. Characterization of carotenoids in soil bacteria and investigation of their photodegradation by UVA radiation via resonance Raman spectroscopy. Analyst 2015, 140, 4584–4593. [Google Scholar] [CrossRef]
- Burkart, N.; Huber, M.F. A survey on the explainability of supervised machine learning. J. Artif. Intell. Res. 2021, 70, 245–317. [Google Scholar] [CrossRef]
- Goodwin, N.L.; Nilsson, S.R.O.; Choong, J.J.; Golden, S.A. Toward the explainability, transparency, and universality of machine learning for behavioral classification in neuroscience. Curr. Opin. Neurobiol. 2022, 73, 102544. [Google Scholar] [CrossRef]
- Vinogradova, K.; Dibrov, A.; Myers, G. Towards Interpretable Semantic Segmentation via Gradient-Weighted Class Activation Mapping (Student Abstract). Proc. AAAI Conf. Artif. Intell. 2020, 34, 13943–13944. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Why did you say that? Visual explanations from deep networks via gradient-based localization. Revista do Hospital das Clínicas 2016, 17, 331–336. [Google Scholar]
- Tewes, T.J.; Centeleghe, I.; Maillard, J.-Y.; Platte, F.; Bockmühl, D.P. Raman Microscopic Analysis of Dry-Surface Biofilms on Clinically Relevant Materials. Microorganisms 2022, 10, 1369. [Google Scholar] [CrossRef] [PubMed]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000, 120, 122–125. [Google Scholar]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2, 8. [Google Scholar] [CrossRef]
- Guo, S.; Popp, J.; Bocklitz, T. Chemometric analysis in Raman spectroscopy from experimental design to machine learning–based modeling. Nat. Protoc. 2021, 16, 5426–5459. [Google Scholar] [CrossRef]
- Chang, W.-C. On Using Principal Components Before Separating a Mixture of Two Multivariate Normal Distributions. J. R. Stat. Soc. Ser. C 1983, 32, 267–275. [Google Scholar] [CrossRef]
- De Siqueira e Oliveira, F.S.; Giana, H.E.; Silveira, L. Discrimination of selected species of pathogenic bacteria using near-infrared Raman spectroscopy and principal components analysis. J. Biomed. Opt. 2012, 17, 107004. [Google Scholar] [CrossRef]
- Huang, Z.; Lui, H.; Chen, M.X.; Alajlan, A.; McLean, D.I.; Zeng, H. Raman spectroscopy of in vivo cutaneous melanin. J. Biomed. Opt. 2004, 9, 1198–1205. [Google Scholar] [CrossRef]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Schuster, K.C.; Reese, I.; Urlaub, E.; Gapes, J.R.; Lendl, B. Multidimensional Information on the Chemical Composition of Single Bacterial Cells by Confocal Raman Microspectroscopy. Anal. Chem. 2000, 72, 5529–5534. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]



| Microorganism | Abbreviation | Number of Spectra | |
|---|---|---|---|
| Fungal spores | Metarhizium brunneum Cb16III | Cb16III | 642 |
| Metarhizium brunneum Ca8II | Ca8II | 562 | |
| Metarhizium brunneum Cb15III | Cb15III | 525 | |
| Metarhizium pemphigi X1c | Mpemp | 847 | |
| Beauveria bassiana | Bbass | 372 | |
| Carotenoid- containing | Chryseobacterium indolgenes | Cin | 684 |
| Kocuria rosea | Kro | 639 | |
| Micrococcus luteus | Mlu | 1842 | |
| Staphylococcus aureus | Sau | 1094 | |
| Xanthophyllomyces dendrorhous | Xde | 658 |
| Microorganism | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
| Cb16III | 0.98 | 0.97 | 0.97 | 131 |
| Ca8II | 0.88 | 0.83 | 0.86 | 133 |
| Cb15 | 0.96 | 0.97 | 0.96 | 105 |
| Mpemp | 0.89 | 0.92 | 0.90 | 181 |
| Bbass | 1.00 | 1.00 | 0.99 | 74 |
| accuracy | 0.93 | 624 | ||
| macro avg | 0.94 | 0.93 | 0.94 | 624 |
| weighted avg | 0.93 | 0.93 | 0.93 | 624 |
| Microorganism | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
| Cb16III | 0.97 | 0.98 | 0.98 | 131 |
| Ca8II | 0.90 | 0.82 | 0.85 | 133 |
| Cb15 | 0.98 | 0.97 | 0.97 | 105 |
| Mpemp | 0.87 | 0.93 | 0.90 | 181 |
| Bbass | 1.00 | 1.00 | 0.99 | 74 |
| accuracy | 0.93 | 624 | ||
| macro avg | 0.94 | 0.94 | 0.94 | 624 |
| weighted avg | 0.93 | 0.93 | 0.93 | 624 |
| Microorganism | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
| Cin | 1.00 | 1.00 | 1.00 | 137 |
| Kro | 1.00 | 1.00 | 1.00 | 128 |
| Mlu | 1.00 | 1.00 | 1.00 | 368 |
| Sau | 1.00 | 1.00 | 1.00 | 219 |
| Xde | 1.00 | 1.00 | 1.00 | 130 |
| accuracy | 1.00 | 983 | ||
| macro avg | 1.00 | 1.00 | 1.00 | 983 |
| weighted avg | 1.00 | 1.00 | 1.00 | 983 |
| Microorganism | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
| Cin | 1.00 | 1.00 | 1.00 | 137 |
| Kro | 1.00 | 1.00 | 1.00 | 128 |
| Mlu | 1.00 | 1.00 | 1.00 | 368 |
| Sau | 1.00 | 1.00 | 1.00 | 219 |
| Xde | 1.00 | 1.00 | 1.00 | 130 |
| accuracy | 1.00 | 983 | ||
| macro avg | 1.00 | 1.00 | 1.00 | 983 |
| weighted avg | 1.00 | 1.00 | 1.00 | 983 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tewes, T.J.; Welle, M.C.; Hetjens, B.T.; Tipatet, K.S.; Pavlov, S.; Platte, F.; Bockmühl, D.P. Understanding Raman Spectral Based Classifications with Convolutional Neural Networks Using Practical Examples of Fungal Spores and Carotenoid-Pigmented Microorganisms. AI 2023, 4, 114-127. https://doi.org/10.3390/ai4010006
Tewes TJ, Welle MC, Hetjens BT, Tipatet KS, Pavlov S, Platte F, Bockmühl DP. Understanding Raman Spectral Based Classifications with Convolutional Neural Networks Using Practical Examples of Fungal Spores and Carotenoid-Pigmented Microorganisms. AI. 2023; 4(1):114-127. https://doi.org/10.3390/ai4010006
Chicago/Turabian StyleTewes, Thomas J., Michael C. Welle, Bernd T. Hetjens, Kevin Saruni Tipatet, Svyatoslav Pavlov, Frank Platte, and Dirk P. Bockmühl. 2023. "Understanding Raman Spectral Based Classifications with Convolutional Neural Networks Using Practical Examples of Fungal Spores and Carotenoid-Pigmented Microorganisms" AI 4, no. 1: 114-127. https://doi.org/10.3390/ai4010006
APA StyleTewes, T. J., Welle, M. C., Hetjens, B. T., Tipatet, K. S., Pavlov, S., Platte, F., & Bockmühl, D. P. (2023). Understanding Raman Spectral Based Classifications with Convolutional Neural Networks Using Practical Examples of Fungal Spores and Carotenoid-Pigmented Microorganisms. AI, 4(1), 114-127. https://doi.org/10.3390/ai4010006






