Influence of Inorganic Nutrients on a North Atlantic Microbial Community’s Response to Ocean Alkalinity Enhancement
Abstract
1. Introduction
2. Materials and Methods
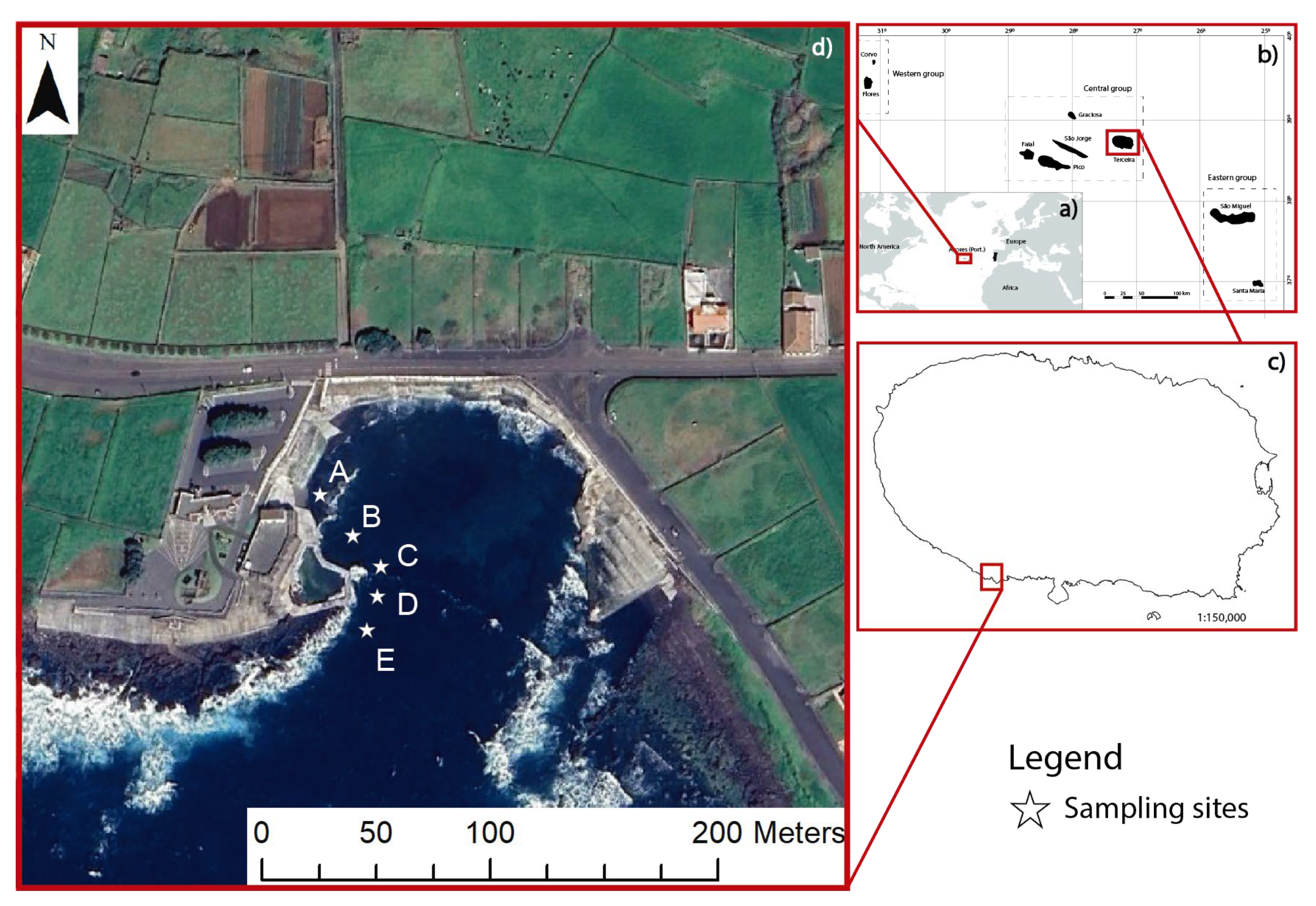
2.1. Sampling Site
2.2. Experimental Setup
2.3. Parameters Evaluated
2.4. Data Analysis
3. Results
3.1. Carbonate Chemistry and Nutrient Conditions
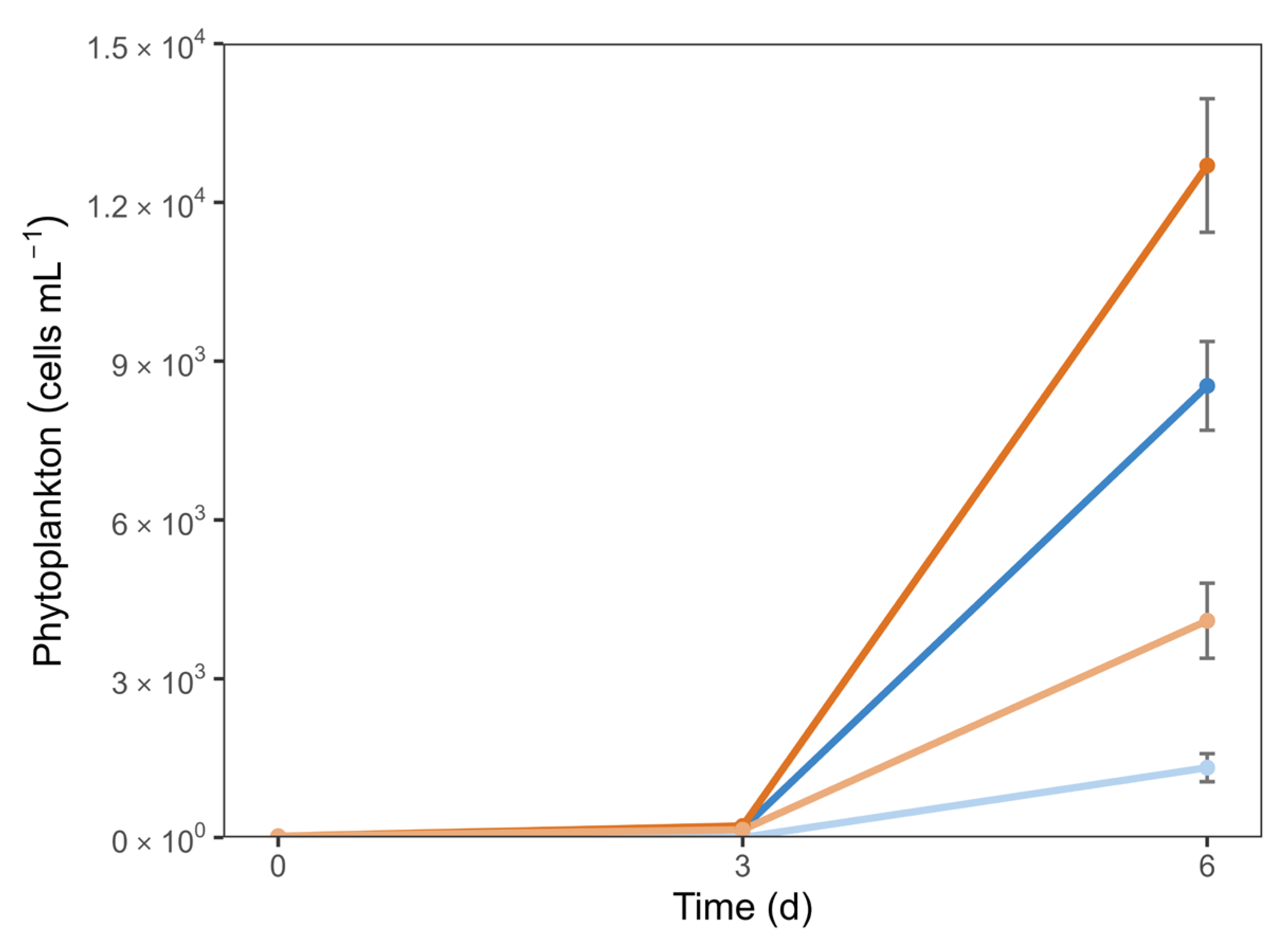
3.2. Phytoplankton Dynamics
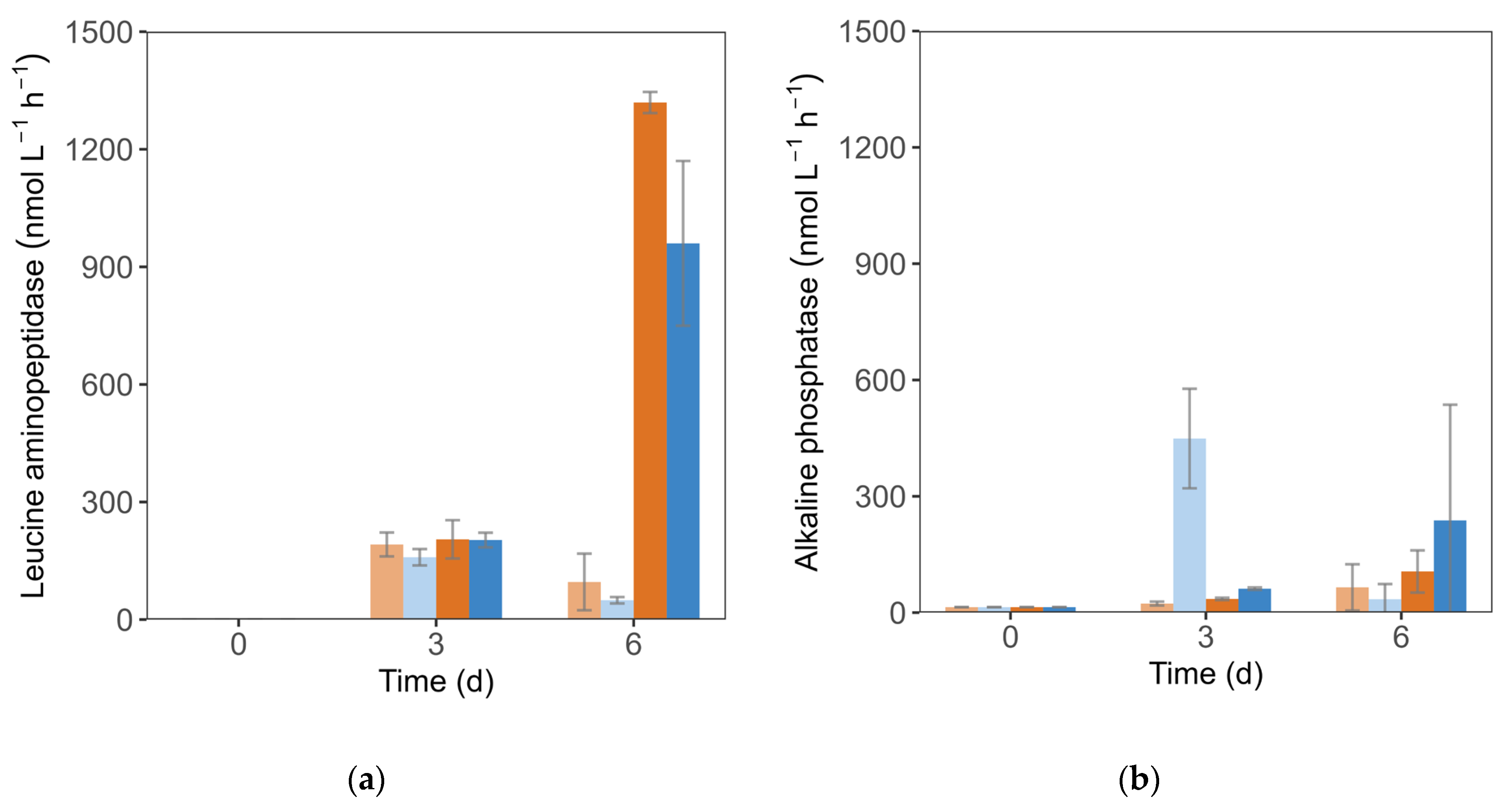
3.3. Enzymatic Activity
3.4. Bacterioplankton Community Structure
4. Discussion
4.1. Drivers of Phytoplankton Response to Ocean Alkalinity Enhancement
4.2. Phytoplankton-Mediated Enzymatic Activity Under Ocean Alkalinity Enhancement
4.3. Phytoplankton–Bacteria Coupling Shapes Microbial Responses to OAE
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| CCM | Carbon-concentrating mechanism |
| LAP | Leucine aminopeptidase |
| OAE | Ocean Alkalinity Enhancement |
| TA | Total Alkalinity |
References
- United Nations Framework Convention on Climate Change Paris Agreement; United Nations: New York, NY, USA, 2015.
- Calvin, K.; Dasgupta, D.; Krinner, G.; Mukherji, A.; Thorne, P.W.; Trisos, C.; Romero, J.; Aldunce, P.; Barrett, K.; Blanco, G.; et al. IPCC, 2023: Climate Change 2023: Synthesis Report, Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Lee, H., Romero, J., Eds.; IPCC: Geneva, Switzerland, 2023. [Google Scholar]
- Low, S.; Baum, C.M.; Sovacool, B.K. Rethinking Net-Zero Systems, Spaces, and Societies: “Hard” versus “Soft” Alternatives for Nature-Based and Engineered Carbon Removal. Glob. Environ. Change 2022, 75, 102530. [Google Scholar] [CrossRef]
- Rau, G.H.; McLeod, E.L.; Hoegh-Guldberg, O. The Need for New Ocean Conservation Strategies in a High-Carbon Dioxide World. Nat. Clim. Chang. 2012, 2, 720–724. [Google Scholar] [CrossRef]
- Bach, L.T.; Gill, S.J.; Rickaby, R.E.M.; Gore, S.; Renforth, P. CO2 Removal with Enhanced Weathering and Ocean Alkalinity Enhancement: Potential Risks and Co-Benefits for Marine Pelagic Ecosystems. Front. Clim. 2019, 1, 7. [Google Scholar] [CrossRef]
- Gattuso, J.-P.; Williamson, P.; Duarte, C.M.; Magnan, A.K. The Potential for Ocean-Based Climate Action: Negative Emissions Technologies and Beyond. Front. Clim. 2021, 2, 575716. [Google Scholar] [CrossRef]
- Hartmann, J.; West, A.J.; Renforth, P.; Köhler, P.; De La Rocha, C.L.; Wolf-Gladrow, D.A.; Dürr, H.H.; Scheffran, J. Enhanced Chemical Weathering as a Geoengineering Strategy to Reduce Atmospheric Carbon Dioxide, Supply Nutrients, and Mitigate Ocean Acidification. Rev. Geophys. 2013, 51, 113–149. [Google Scholar] [CrossRef]
- Kheshgi, H.S. Sequestering Atmospheric Carbon Dioxide by Increasing Ocean Alkalinity. Energy 1995, 20, 915–922. [Google Scholar] [CrossRef]
- Renforth, P.; Henderson, G. Assessing Ocean Alkalinity for Carbon Sequestration. Rev. Geophys. 2017, 55, 636–674. [Google Scholar] [CrossRef]
- Feng, E.Y.; Keller, D.P.; Koeve, W.; Oschlies, A. Could Artificial Ocean Alkalinization Protect Tropical Coral Ecosystems from Ocean Acidification? Environ. Res. Lett. 2016, 11, 074008. [Google Scholar] [CrossRef]
- Gattuso, J.P.; Magnan, A.K.; Bopp, L.; Cheung, W.W.L.; Duarte, C.M.; Hinkel, J.; Mcleod, E.; Micheli, F.; Oschlies, A.; Williamson, P.; et al. Ocean Solutions to Address Climate Change and Its Effects on Marine Ecosystems. Front. Mar. Sci. 2018, 5, 337. [Google Scholar] [CrossRef]
- Keller, D.P.; Feng, E.Y.; Oschlies, A. Potential Climate Engineering Effectiveness and Side Effects during a High Carbon Dioxide-Emission Scenario. Nat. Commun. 2014, 5, 3304. [Google Scholar] [CrossRef]
- Eisaman, M.D.; Geilert, S.; Renforth, P.; Bastianini, L.; Campbell, J.; Dale, A.W.; Foteinis, S.; Grasse, P.; Hawrot, O.; Löscher, C.R.; et al. Assessing the Technical Aspects of Ocean-Alkalinity-Enhancement Approaches. State Planet. 2023, 3, 2-oae2023. [Google Scholar] [CrossRef]
- Foteinis, S.; Andresen, J.; Campo, F.; Caserini, S.; Renforth, P. Life Cycle Assessment of Ocean Liming for Carbon Dioxide Removal from the Atmosphere. J. Clean. Prod. 2022, 370, 133309. [Google Scholar] [CrossRef]
- Renforth, P.; Jenkins, B.G.; Kruger, T. Engineering Challenges of Ocean Liming. Energy 2013, 60, 442–452. [Google Scholar] [CrossRef]
- Marín-Samper, L.; Arístegui, J.; Hernández-Hernández, N.; Ortiz, J.; Archer, S.D.; Ludwig, A.; Riebesell, U. Assessing the Impact of CO2-Equilibrated Ocean Alkalinity Enhancement on Microbial Metabolic Rates in an Oligotrophic System. Biogeosciences 2024, 21, 2859–2876. [Google Scholar] [CrossRef]
- Ramírez, L.; Pozzo-Pirotta, L.J.; Trebec, A.; Manzanares-Vázquez, V.; Díez, J.L.; Arístegui, J.; Riebesell, U.; Archer, S.D.; Segovia, M. Ocean Alkalinity Enhancement (OAE) Does Not Cause Cellular Stress in a Phytoplankton Community of the Subtropical Atlantic Ocean. Biogeosciences 2025, 22, 1865–1886. [Google Scholar] [CrossRef]
- Subhas, A.V.; Marx, L.; Reynolds, S.; Flohr, A.; Mawji, E.W.; Brown, P.J.; Cael, B.B. Microbial Ecosystem Responses to Alkalinity Enhancement in the North Atlantic Subtropical Gyre. Front. Clim. 2022, 4, 784997. [Google Scholar] [CrossRef]
- Suessle, P.; Taucher, J.; Goldenberg, S.U.; Baumann, M.; Spilling, K.; Noche-Ferreira, A.; Vanharanta, M.; Riebesell, U. Particle Fluxes by Subtropical Pelagic Communities under Ocean Alkalinity Enhancement. Biogeosciences 2025, 22, 71–86. [Google Scholar] [CrossRef]
- Guo, J.A.; Strzepek, R.F.; Swadling, K.M.; Townsend, A.T.; Bach, L.T. Influence of Ocean Alkalinity Enhancement with Olivine or Steel Slag on a Coastal Plankton Community in Tasmania. Biogeosciences 2024, 21, 2335–2354. [Google Scholar] [CrossRef]
- Guo, J.A.; Strzepek, R.F.; Yuan, Z.; Swadling, K.M.; Townsend, A.T.; Achterberg, E.P.; Browning, T.J.; Bach, L.T. Effects of Ocean Alkalinity Enhancement on Plankton in the Equatorial Pacific. Commun. Earth Env. 2025, 6, 270. [Google Scholar] [CrossRef]
- Ferderer, A.; Chase, Z.; Kennedy, F.; Schulz, K.G.; Bach, L.T. Assessing the Influence of Ocean Alkalinity Enhancement on a Coastal Phytoplankton Community. Biogeosciences 2022, 19, 5375–5399. [Google Scholar] [CrossRef]
- Xin, X.; Goldenberg, S.U.; Taucher, J.; Stuhr, A.; Arístegui, J.; Riebesell, U. Resilience of Phytoplankton and Microzooplankton Communities under Ocean Alkalinity Enhancement in the Oligotrophic Ocean. Env. Sci. Technol. 2024, 58, 20918–20930. [Google Scholar] [CrossRef]
- Barcelos e Ramos, J.; Schulz, K.G.; Voss, M.; Narciso, Á.; Müller, M.N.; Reis, F.V.; Cachão, M.; Azevedo, E.B. Nutrient-Specific Responses of a Phytoplankton Community: A Case Study of the North Atlantic Gyre, Azores. J. Plankton Res. 2017, 39, 744–761. [Google Scholar] [CrossRef]
- Crummett, L.T. Acidification Decreases Microbial Community Diversity in the Salish Sea, a Region with Naturally High pCO2. PLoS ONE 2020, 15, e0241183. [Google Scholar] [CrossRef] [PubMed]
- Lin, S. Phosphate Limitation and Ocean Acidification Co-Shape Phytoplankton Physiology and Community Structure. Nat. Commun. 2023, 14, 2699. [Google Scholar] [CrossRef] [PubMed]
- Burt, D.J.; Fröb, F.; Ilyina, T. The Sensitivity of the Marine Carbonate System to Regional Ocean Alkalinity Enhancement. Front. Clim. 2021, 3, 624075. [Google Scholar] [CrossRef]
- Jürchott, M.; Oschlies, A.; Koeve, W. Artificial Upwelling—A Refined Narrative. Geophys. Res. Lett. 2023, 50, e2022GL101870. [Google Scholar] [CrossRef]
- Moras, C.A.; Bach, L.T.; Cyronak, T.; Joannes-Boyau, R.; Schulz, K.G. Ocean Alkalinity Enhancement—Avoiding Runaway CaCO3 Precipitation during Quick and Hydrated Lime Dissolution. Biogeosciences 2022, 19, 3537–3557. [Google Scholar] [CrossRef]
- Dickson, A.G.; Afghan, J.D.; Anderson, G.C. Reference Materials for Oceanic CO2 Analysis: A Method for the Certification of Total Alkalinity. Mar. Chem. 2003, 80, 185–197. [Google Scholar] [CrossRef]
- Lewis, E.; Wallace, D.; Allison, L.J. Program Developed for CO2 System Calculations; Oak Ridge National Laboratory: Oak Ridge, TN, USA, 1998. [Google Scholar]
- Lueker, T.J.; Dickson, A.G.; Keeling, C.D. Ocean PCO Calculated from Dissolved Inorganic Carbon, 2 Alkalinity, and Equations for K and K: Validation Based on 1 2 Laboratory Measurements of CO in Gas and Seawater at 2 Equilibrium. Mar. Chem. 2000, 70, 105–119. [Google Scholar] [CrossRef]
- Hansen, H.; Koroleff, F. Determination of Nutrients. In Methods of Seawater Analysis; Wiley: Hoboken, NJ, USA, 1999. [Google Scholar]
- Hoppe, H.-G. Significance of Exoenzymatic Activities in the Ecology of Brackish Water: Measurements by Means of Methylumbelliferyl-Substrates. Mar. Ecol. Prog. Ser. 1983, 11, 299–308. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Posit Team. RStudio: Integrated Development Environment for R, Version 2025.05.0; Posit Software PBC: Boston, MA, USA, 2025.
- Reynolds, C.S. The Response of Phytoplankton Communities to Changing Lake Environments. Swiss J. Hydrol. 1987, 49, 220–236. [Google Scholar] [CrossRef]
- de Castro, I.; Ribeiro, S.C.; Louvado, A.; Gomes, N.C.M.; Cachão, M.; Brito de Azevedo, E.; Barcelos e Ramos, J. Ocean Liming Effect on a North Atlantic Microbial Community: Changes in Composition and Rates. Front. Mar. Sci. 2025, 12, 1602158. [Google Scholar] [CrossRef]
- Marín-Samper, L.; Arístegui, J.; Hernández-Hernández, N.; Riebesell, U. Responses of Microbial Metabolic Rates to Non-Equilibrated Silicate- versus Calcium-Based Ocean Alkalinity Enhancement. Biogeosciences 2024, 21, 5707–5724. [Google Scholar] [CrossRef]
- Matsuda, Y.; Hopkinson, B.M.; Nakajima, K.; Dupont, C.L.; Tsuji, Y. Mechanisms of Carbon Dioxide Acquisition and CO 2 Sensing in Marine Diatoms: A Gateway to Carbon Metabolism. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160403. [Google Scholar] [CrossRef]
- Hopkinson, B.M.; Dupont, C.L.; Matsuda, Y. The Physiology and Genetics of CO2 Concentrating Mechanisms in Model Diatoms. Curr. Opin. Plant Biol. 2016, 31, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Oberlander, J.L.; Burke, M.E.; London, C.A.; Macintyre, H.L. Assessing the Impacts of Simulated Ocean Alkalinity Enhancement on Viability and Growth of Nearshore Species of Phytoplankton. Biogeosciences 2025, 22, 499–512. [Google Scholar] [CrossRef]
- Gately, J.A.; Kim, S.M.; Jin, B.; Brzezinski, M.A.; Iglesias-Rodriguez, M.D. Coccolithophores and Diatoms Resilient to Ocean Alkalinity Enhancement: A Glimpse of Hope? Sci. Adv. 2023, 9, adg6066. [Google Scholar] [CrossRef]
- Shi, Z.; Xu, J.; Li, X.; Li, R.; Li, Q. Links of Extracellular Enzyme Activities, Microbial Metabolism, and Community Composition in the River-Impacted Coastal Waters. J. Geophys. Res. Biogeosci 2019, 124, 3507–3520. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master Recyclers: Features and Functions of Bacteria Associated with Phytoplankton Blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef]
- McIlroy, S.J.; Nielsen, P.H. The Family Saprospiraceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2014; pp. 863–889. [Google Scholar]
- Xia, Y.; Kong, Y.; Thomsen, T.R.; Halkjær Nielsen, P. Identification and Ecophysiological Characterization of Epiphytic Protein-Hydrolyzing Saprospiraceae (“Candidatus Epiflobacter” spp.) in Activated Sludge. Appl. Environ. Microbiol. 2008, 74, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, H.-G. Phosphatase Activity in the Sea. Hydrobiologia 2003, 493, 187–200. [Google Scholar] [CrossRef]
- Korlević, M.; Markovski, M.; Zhao, Z.; Herndl, G.J.; Najdek, M. Seasonal Dynamics of Epiphytic Microbial Communities on Marine Macrophyte Surfaces. Front. Microbiol. 2021, 12, 671342. [Google Scholar] [CrossRef]
- Miranda, L.N.; Hutchison, K.; Grossman, A.R.; Brawley, S.H. Diversity and Abundance of the Bacterial Community of the Red Macroalga Porphyra Umbilicalis: Did Bacterial Farmers Produce Macroalgae? PLoS ONE 2013, 8, e58269. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.; Levold, F.; Allgaier, M.; Simon, M.; Brinkhoff, T. Marine Diatom Species Harbour Distinct Bacterial Communities. Env. Microbiol. 2005, 7, 860–873. [Google Scholar] [CrossRef]
- Pontiller, B.; Martínez-García, S.; Joglar, V.; Amnebrink, D.; Pérez-Martínez, C.; González, J.M.; Lundin, D.; Fernández, E.; Teira, E.; Pinhassi, J. Rapid Bacterioplankton Transcription Cascades Regulate Organic Matter Utilization during Phytoplankton Bloom Progression in a Coastal Upwelling System. ISME J. 2022, 16, 2360–2372. [Google Scholar] [CrossRef]
- Delgadillo-Nuño, E.; Teira, E.; Pontiller, B.; Lundin, D.; Joglar, V.; Pedrós-Alió, C.; Fernández, E.; Pinhassi, J.; Martínez-García, S. Coastal Upwelling Systems as Dynamic Mosaics of Bacterioplankton Functional Specialization. Front. Mar. Sci. 2023, 10, 1259783. [Google Scholar] [CrossRef]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-Controlled Succession of Marine Bacterioplankton Populations Induced by a Phytoplankton Bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between Diatoms and Bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef]
- Eigemann, F.; Rahav, E.; Grossart, H.; Aharonovich, D.; Sher, D.; Vogts, A.; Voss, M. Phytoplankton Exudates Provide Full Nutrition to a Subset of Accompanying Heterotrophic Bacteria via Carbon, Nitrogen and Phosphorus Allocation. Environ. Microbiol. 2022, 24, 2467–2483. [Google Scholar] [CrossRef]
- Kieft, B.; Li, Z.; Bryson, S.; Hettich, R.L.; Pan, C.; Mayali, X.; Mueller, R.S. Phytoplankton Exudates and Lysates Support Distinct Microbial Consortia with Specialized Metabolic and Ecophysiological Traits. Proc. Natl. Acad. Sci. USA 2021, 118, e2101178118. [Google Scholar] [CrossRef]
- Seymour, J.R.; Amin, S.A.; Raina, J.-B.; Stocker, R. Zooming in on the Phycosphere: The Ecological Interface for Phytoplankton–Bacteria Relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef]
- Bunse, C.; Lundin, D.; Karlsson, C.M.G.; Akram, N.; Vila-Costa, M.; Palovaara, J.; Svensson, L.; Holmfeldt, K.; González, J.M.; Calvo, E.; et al. Response of Marine Bacterioplankton PH Homeostasis Gene Expression to Elevated CO2. Nat. Clim. Change 2016, 6, 483–487. [Google Scholar] [CrossRef]
- Huang, R.; Sun, J.; Yang, Y.; Jiang, X.; Wang, Z.; Song, X.; Wang, T.; Zhang, D.; Li, H.; Yi, X.; et al. Elevated pCO2 Impedes Succession of Phytoplankton Community from Diatoms to Dinoflagellates Along with Increased Abundance of Viruses and Bacteria. Front. Mar. Sci. 2021, 8, 642208. [Google Scholar] [CrossRef]
- Sperling, M.; Piontek, J.; Gerdts, G.; Wichels, A.; Schunck, H.; Roy, A.-S.; La Roche, J.; Gilbert, J.; Nissimov, J.I.; Bittner, L.; et al. Effect of Elevated CO2 on the Dynamics of Particle-Attached and Free-Living Bacterioplankton Communities in an Arctic Fjord. Biogeosciences 2013, 10, 181–191. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Yang, Y.; Tu, Q.; Zhou, J.; Jiao, N. Ocean Acidification Altered Microbial Functional Potential in the Arctic Ocean. Limnol. Ocean. 2023, 68, S217–S229. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Yang, Y.; Luo, T.; Van Nostrand, J.D.; Zhou, J.; Jiao, N.; Zhang, R. Ocean Acidification Regulates the Activity, Community Structure, and Functional Potential of Heterotrophic Bacterioplankton in an Oligotrophic Gyre. J. Geophys. Res. Biogeosci. 2019, 124, 1001–1017. [Google Scholar] [CrossRef]



| Nutrient Condition | NO3− (µM) | PO43− (µM) | Si(OH)4 (µM) | NO2− (µM) |
|---|---|---|---|---|
| NN | 7.91 | 0.24 | 47.81 | 0.31 |
| EN | 80.77 | 3.48 | 44.38 | 0.17 |
| TA (µmol kg−1) | pHt | pCO2 (µatm) | HCO3− (µmol kg−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nutrients | Alkalinity | T0 | T3 | T6 | T0 | T3 | T6 | T0 | T3 | T6 | T0 | T3 | T6 |
| NN | Ambient | 2353 (0) | 2344 (0.89) | 2359 (1.78) | 8.05 (0) | 8.05 (0) | 8.28 (0.06) | 409 (0) | 399 (2.36) | 219 (37.99) | 1915 (0) | 1901 (5.82) | 1696 (2.33) |
| OAE | 2530 (0) | 2516 (1.39) | 2528 (0.74) | 8.28 (0) | 8.22 (0) | 8.25 (0) | 227 (0) | 267 (0.56) | 253 (3.30) | 1824 (0) | 1880 (1.65) | 1865 (4.75) | |
| EN | Ambient | 2353 (0) | 2332 (2.25) | 2406 (3.09) | 8.03 (0) | 8.01 (0) | 8.68 (0.03) | 429 (0) | 445 (1.65) | 60 (5.36) | 1928 (0) | 1923 (0.89) | 1211 (38.53) |
| OAE | 2524 (0) | 2505 (1.73) | 2537 (7.20) | 8.28 (0) | 8.24 (0.01) | 8.71 (0) | 228 (0) | 257 (3.73) | 57 (0.18) | 1820 (0) | 1855 (2.35) | 1243 (59.47) | |
| Orders | NN p-Value | EN p-Value | Effect of OAE |
|---|---|---|---|
| Oceanospirillales | 0.229 | 0.798 | − |
| Flavobacteriales | 0.764 | 0.002 | ↓ under EN |
| Vibrionales | 0.918 | − | − |
| Chitinophagales | 0.018 | 0.007 | ↓ under NN and EN |
| Alteromonadales | 0.718 | 0.528 | − |
| Cellvibrionales | 0.026 | − | ↑ under NN |
| Rhodobacterales | 0.194 | 0.0391 | ↓ under EN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Castro, I.; Ribeiro, S.C.; Louvado, A.; Gomes, N.C.M.; Cachão, M.; Silva Borges, P.F.; Brito de Azevedo, E.; Barcelos e Ramos, J. Influence of Inorganic Nutrients on a North Atlantic Microbial Community’s Response to Ocean Alkalinity Enhancement. Oceans 2025, 6, 65. https://doi.org/10.3390/oceans6040065
de Castro I, Ribeiro SC, Louvado A, Gomes NCM, Cachão M, Silva Borges PF, Brito de Azevedo E, Barcelos e Ramos J. Influence of Inorganic Nutrients on a North Atlantic Microbial Community’s Response to Ocean Alkalinity Enhancement. Oceans. 2025; 6(4):65. https://doi.org/10.3390/oceans6040065
Chicago/Turabian Stylede Castro, Inês, Susana C. Ribeiro, António Louvado, Newton Carlos Marcial Gomes, Mário Cachão, Paulo F. Silva Borges, Eduardo Brito de Azevedo, and Joana Barcelos e Ramos. 2025. "Influence of Inorganic Nutrients on a North Atlantic Microbial Community’s Response to Ocean Alkalinity Enhancement" Oceans 6, no. 4: 65. https://doi.org/10.3390/oceans6040065
APA Stylede Castro, I., Ribeiro, S. C., Louvado, A., Gomes, N. C. M., Cachão, M., Silva Borges, P. F., Brito de Azevedo, E., & Barcelos e Ramos, J. (2025). Influence of Inorganic Nutrients on a North Atlantic Microbial Community’s Response to Ocean Alkalinity Enhancement. Oceans, 6(4), 65. https://doi.org/10.3390/oceans6040065








