Research on Obtaining Pepper Phenotypic Parameters Based on Improved YOLOX Algorithm
Abstract
1. Introduction
- Construction of the Pepper-mini dataset. This study verifies the conversion relationship of unit pixels at different distances for the detection system. It utilizes offline data augmentation to construct the Pepper-mini dataset from the collected pepper image dataset, which can be openly used.
- Optimization of YOLOX-tiny object detection algorithm using the CA attention mechanism. This study enhances the network’s feature extraction capability for peppers, improving the overall object detection performance. It identifies target plants based on the Euclidean distance-based object detection box filtering rule. This study utilizes image processing operations such as threshold segmentation, morphological changes, and connected area denoising in the HSV color space to extract binary images of the processed target plants. Finally, this study applies plant height and stem diameter measurement algorithms to achieve precise measurements of the target plants.
- Proposal and application of a novel plant height and stem diameter measurement algorithm, which to some extent can replace manual measurements.
2. Materials and Methods
2.1. Construction of the Pepper-Mini Dataset
2.1.1. Data Collection
2.1.2. Data Augmentation
- Dataset with rotation, flipping, and random cropping. The initial dataset was rotated by 10° and 350°, and horizontally flipped.
- Dataset with brightness adjustment. The initial dataset was transformed from the RGB color space to the HSV color space, and then the brightness (Value, V) channel was increased by 10% and decreased by 10%.
- Dataset with added noise. The initial dataset was processed with salt-and-pepper noise and Gaussian noise.
2.1.3. Data Annotation
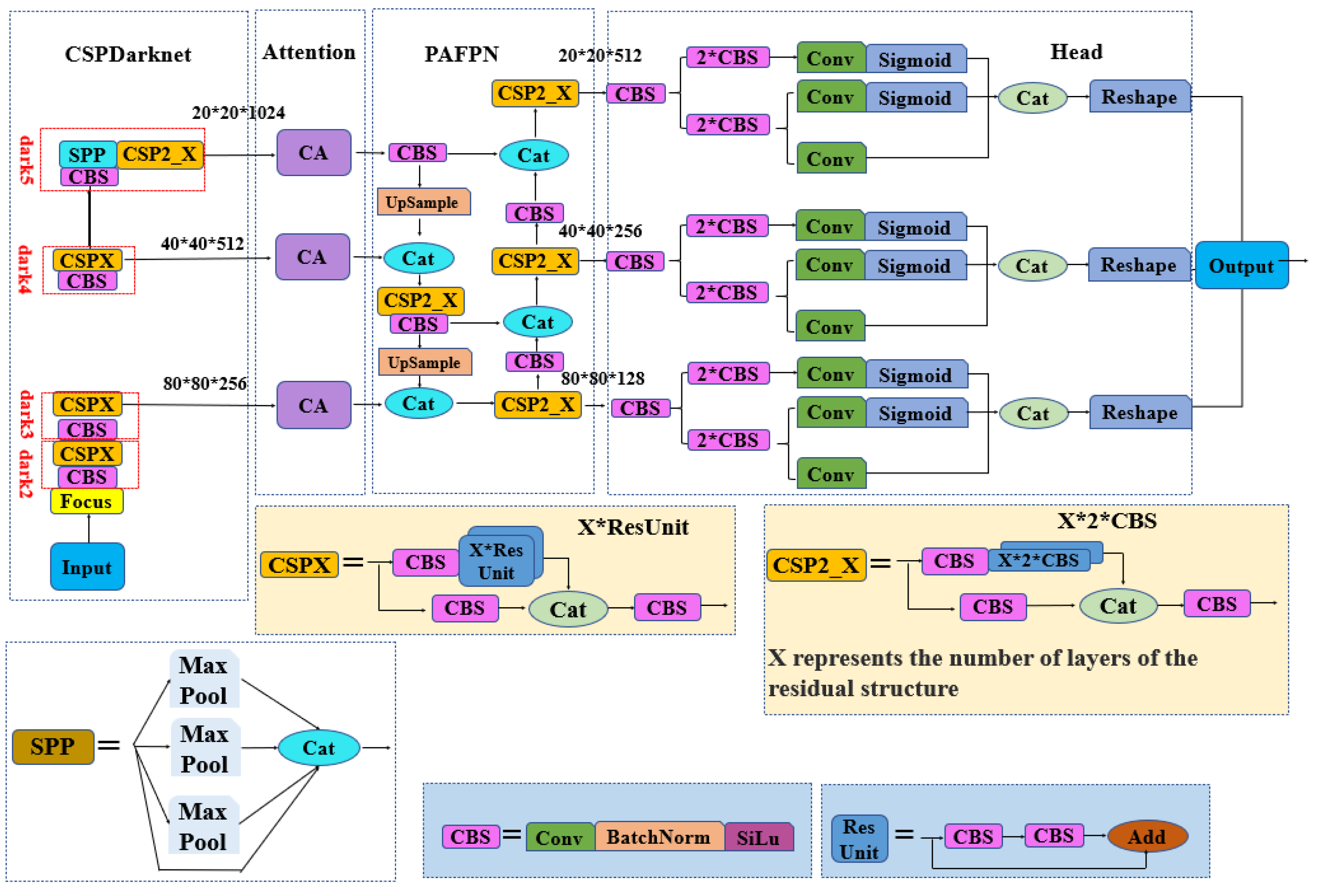
2.2. Improving the YOLOX Model Based on the Channel Attention (CA) Mechanism
2.2.1. YOLOX Model
2.2.2. Coordinate Attention Mechanism
2.2.3. The YOLOX Model Integrated with the Coordinate Attention (CA) Mechanism
2.2.4. Model Training Parameter Settings and Evaluation Metrics
2.3. Obtaining Pepper Phenotypic Parameters
2.3.1. Phenotypic Parameter Extraction: Image Preprocessing
2.3.2. Plant Height and Stem Thickness Measurement Algorithm
3. Experiment and Result Analysis
3.1. Model Comparison
3.2. Plant Height Stem Diameter Measurement
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dobón-Suárez, A.; Zapata, P.J.; García-Pastor, M.E. A Comprehensive Review on Characterization of Pepper Seeds: Unveiling Potential Value and Sustainable Agrifood Applications. Foods 2025, 14, 1969. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, S.; Pabitha, C.; Kumar, T.R.; Joseph, J.; Mahendran, G.; Maranan, R. Enhancing pepper growth and yield through disease identification in plants using leaf-based deep learning techniques. In Hybrid and Advanced Technologies; CRC Press: Boca Raton, FL, USA, 2025; pp. 54–59. [Google Scholar]
- Gerakari, M.; Katsileros, A.; Kleftogianni, K.; Tani, E.; Bebeli, P.J.; Papasotiropoulos, V. Breeding of Solanaceous Crops Using AI: Machine Learning and Deep Learning Approaches—A Critical Review. Agronomy 2025, 15, 757. [Google Scholar] [CrossRef]
- Sanatombi, K. A comprehensive review on sustainable strategies for valorization of pepper waste and their potential application. Compr. Rev. Food Sci. Food Saf. 2025, 24, e70118. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X.; Yang, W.; Peng, G.; Chen, J.; Yin, Y.; Yan, J. An efficient method for chili pepper variety classification and origin tracing based on an electronic nose and deep learning. Food Chem. 2025, 479, 143850. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, S. YOLOv8-CBSE: An Enhanced Computer Vision Model for Detecting the Maturity of Chili Pepper in the Natural Environment. Agronomy 2025, 15, 537. [Google Scholar] [CrossRef]
- Wang, R.F.; Su, W.H. The application of deep learning in the whole potato production Chain: A Comprehensive review. Agriculture 2024, 14, 1225. [Google Scholar] [CrossRef]
- Jiang, L.; Rodriguez-Sanchez, J.; Snider, J.L.; Chee, P.W.; Fu, L.; Li, C. Mapping of cotton bolls and branches with high-granularity through point cloud segmentation. Plant Methods 2025, 21, 1–24. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Xia, W.K.; Chu, H.Q.; Su, W.H.; Wang, R.F.; Wang, H. A Comprehensive Review of Deep Learning Applications in Cotton Industry: From Field Monitoring to Smart Processing. Plants 2025, 14, 1481. [Google Scholar] [CrossRef]
- Jiang, L.; Li, C.; Fu, L. Apple tree architectural trait phenotyping with organ-level instance segmentation from point cloud. Comput. Electron. Agric. 2025, 229, 109708. [Google Scholar] [CrossRef]
- Fatchurrahman, D.; Hilaili, M.; Russo, L.; Jahari, M.B.; Fathi-Najafabadi, A. Utilizing RGB imaging and machine learning for freshness level determination of green bell pepper (Capsicum annuum L.) throughout its shelf-life. Postharvest Biol. Technol. 2025, 222, 113359. [Google Scholar] [CrossRef]
- Zheng, X.; Shao, Z.; Chen, Y.; Zeng, H.; Chen, J. MSPB-YOLO: High-Precision Detection Algorithm of Multi-Site Pepper Blight Disease Based on Improved YOLOv8. Agronomy 2025, 15, 839. [Google Scholar] [CrossRef]
- Qin, Y.M.; Tu, Y.H.; Li, T.; Ni, Y.; Wang, R.F.; Wang, H. Deep Learning for Sustainable Agriculture: A Systematic Review on Applications in Lettuce Cultivation. Sustainability 2025, 17, 3190. [Google Scholar] [CrossRef]
- Tan, C.; Sun, J.; Song, H.; Li, C. A customized density map model and segment anything model for cotton boll number, size, and yield prediction in aerial images. Comput. Electron. Agric. 2025, 232, 110065. [Google Scholar] [CrossRef]
- Meyer, G.E.; Davison, D.A. An electronic image plant growth measurement system. Trans. ASAE 1987, 30, 242–0248. [Google Scholar] [CrossRef]
- Casady, W.; Singh, N.; Costello, T. Machine vision for measurement of rice canopy dimensions. Trans. ASAE 1996, 39, 1891–1898. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, R.; Wang, M.; Lai, T.; Zhang, M. Self-supervised transformer-based pre-training method with General Plant Infection dataset. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Urumqi, China, 18–20 October 2024; Springer: Singapore, 2024; pp. 189–202. [Google Scholar]
- Chen, D.; Neumann, K.; Friedel, S.; Kilian, B.; Chen, M.; Altmann, T.; Klukas, C. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 2014, 26, 4636–4655. [Google Scholar] [CrossRef]
- Mora, M.; Avila, F.; Carrasco-Benavides, M.; Maldonado, G.; Olguín-Cáceres, J.; Fuentes, S. Automated computation of leaf area index from fruit trees using improved image processing algorithms applied to canopy cover digital photograpies. Comput. Electron. Agric. 2016, 123, 195–202. [Google Scholar] [CrossRef]
- Zhu, T.; Ma, X.; Guan, H.; Wu, X.; Wang, F.; Yang, C.; Jiang, Q. A calculation method of phenotypic traits based on three-dimensional reconstruction of tomato canopy. Comput. Electron. Agric. 2023, 204, 107515. [Google Scholar] [CrossRef]
- Polk, S.L.; Cui, K.; Chan, A.H.; Coomes, D.A.; Plemmons, R.J.; Murphy, J.M. Unsupervised diffusion and volume maximization-based clustering of hyperspectral images. Remote Sens. 2023, 15, 1053. [Google Scholar] [CrossRef]
- Cui, K.; Li, R.; Polk, S.L.; Lin, Y.; Zhang, H.; Murphy, J.M.; Plemmons, R.J.; Chan, R.H. Superpixel-based and spatially-regularized diffusion learning for unsupervised hyperspectral image clustering. IEEE Trans. Geosci. Remote Sens. 2024, 62, 4405818. [Google Scholar] [CrossRef]
- Chang-Tao, Z.; Rui-Feng, W.; Yu-Hao, T.; Xiao-Xu, P.; Wen-Hao, S. Automatic lettuce weed detection and classification based on optimized convolutional neural networks for robotic weed control. Agronomy 2024, 14, 2838. [Google Scholar] [CrossRef]
- Cui, K.; Tang, W.; Zhu, R.; Wang, M.; Larsen, G.D.; Pauca, V.P.; Alqahtani, S.; Yang, F.; Segurado, D.; Fine, P.; et al. Real-time localization and bimodal point pattern analysis of palms using uav imagery. arXiv 2024, arXiv:2410.11124. [Google Scholar]
- Wang, R.F.; Tu, Y.H.; Chen, Z.Q.; Zhao, C.T.; Su, W.H. A Lettpoint-Yolov11l Based Intelligent Robot for Precision Intra-Row Weeds Control in Lettuce. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=5162748 (accessed on 30 April 2025).
- Zhang, C.B.; Zhong, Y.; Han, K. Mr. DETR: Instructive Multi-Route Training for Detection Transformers. arXiv 2024, arXiv:2412.10028. [Google Scholar]
- Bai, S.; Zhang, M.; Zhou, W.; Huang, S.; Luan, Z.; Wang, D.; Chen, B. Prompt-based distribution alignment for unsupervised domain adaptation. In Proceedings of the AAAI conference on artificial intelligence, Vancouver, BC, Canada, 26–27 February 2024; Volume 38, pp. 729–737. [Google Scholar]
- Akdoğan, C.; Özer, T.; Oğuz, Y. PP-YOLO: Deep learning based detection model to detect apple and cherry trees in orchard based on Histogram and Wavelet preprocessing techniques. Comput. Electron. Agric. 2025, 232, 110052. [Google Scholar] [CrossRef]
- Wu, A.Q.; Li, K.L.; Song, Z.Y.; Lou, X.; Hu, P.; Yang, W.; Wang, R.F. Deep Learning for Sustainable Aquaculture: Opportunities and Challenges. Sustainability 2025, 17, 5084. [Google Scholar] [CrossRef]
- Cui, K.; Zhu, R.; Wang, M.; Tang, W.; Larsen, G.D.; Pauca, V.P.; Alqahtani, S.; Yang, F.; Segurado, D.; Lutz, D.; et al. Detection and Geographic Localization of Natural Objects in the Wild: A Case Study on Palms. arXiv 2025, arXiv:2502.13023. [Google Scholar]
- Yang, B.; Gao, Z.; Gao, Y.; Zhu, Y. Rapid detection and counting of wheat ears in the field using YOLOv4 with attention module. Agronomy 2021, 11, 1202. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. EggplantDet: An efficient lightweight model for eggplant disease detection. Alex. Eng. J. 2025, 115, 308–323. [Google Scholar] [CrossRef]
- Rohith, D.; Saurabh, P.; Bisen, D. An integrated approach to apple leaf disease detection: Leveraging convolutional neural networks for accurate diagnosis. Multimed. Tools Appl. 2025, 1–36. [Google Scholar] [CrossRef]
- Bi, C.; Bi, X.; Liu, J.; Xie, H.; Zhang, S.; Chen, H.; Wang, M.; Shi, L.; Song, S. Identification of maize kernel varieties based on interpretable ensemble algorithms. Front. Plant Sci. 2025, 16, 1511097. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Liu, H.; Wu, Y.; Zhang, L. Towards improved accuracy of UAV-based wheat ears counting: A transfer learning method of the ground-based fully convolutional network. Expert Syst. Appl. 2022, 191, 116226. [Google Scholar] [CrossRef]
- Baweja, H.S.; Parhar, T.; Mirbod, O.; Nuske, S. Stalknet: A deep learning pipeline for high-throughput measurement of plant stalk count and stalk width. In Proceedings of the Field and Service Robotics: Results of the 11th International Conference, Zurich, Switzerland, 12–15 September 2017; Springer: Cham, Switzerland, 2018; pp. 271–284. [Google Scholar]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating biomass of barley using crop surface models (CSMs) derived from UAV-based RGB imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, C.; Tillman, B.; Brown, N. Robotic Plot-Scale Peanut Counting and Yield Estimation Using LoFTR-Based Image Stitching and Improved RT-DETR. In Proceedings of the 2024 ASABE Annual International Meeting, Anaheim, CA, USA, 28–31 July 2024; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2024; p. 1. [Google Scholar]
- Yuan, P.; Qian, S.; Zhai, Z.; FernánMartínez, J.; Xu, H. Study of chrysanthemum image phenotype on-line classification based on transfer learning and bilinear convolutional neural network. Comput. Electron. Agric. 2022, 194, 106679. [Google Scholar] [CrossRef]
- Vit, A.; Shani, G.; Bar-Hillel, A. Length phenotyping with interest point detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Chen, M.; Liao, J.; Zhu, D.; Zhou, H.; Zou, Y.; Zhang, S.; Liu, L. MCC-Net: A class attention-enhanced multi-scale model for internal structure segmentation of rice seedling stem. Comput. Electron. Agric. 2023, 207, 107717. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, C.; Munoz, P.; Takeda, F.; Leme, B. In-field blueberry fruit phenotyping with a MARS-PhenoBot and customized BerryNet. Comput. Electron. Agric. 2025, 232, 110057. [Google Scholar] [CrossRef]
- Wang, B. Zero-exemplar deep continual learning for crop disease recognition: A study of total variation attention regularization in vision transformers. Front. Plant Sci. 2024, 14, 1283055. [Google Scholar] [CrossRef]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. Yolox: Exceeding yolo series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar]
- Li, Y.; Li, J.; Luo, L.; Wang, L.; Zhi, Q. Tomato ripeness and stem recognition based on improved YOLOX. Sci. Rep. 2025, 15, 1924. [Google Scholar] [CrossRef]
- Miao, W.; Shen, J.; Xu, Q.; Hamalainen, T.; Xu, Y.; Cong, F. SpikingYOLOX: Improved YOLOX Object Detection with Fast Fourier Convolution and Spiking Neural Networks. In Proceedings of the AAAI Conference on Artificial Intelligence, Philadelphia, PA, USA, 25 February–4 March 2025; Volume 39, pp. 1465–1473. [Google Scholar]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate attention for efficient mobile network design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13713–13722. [Google Scholar]
- Tan, C.; Sun, J.; Paterson, A.H.; Song, H.; Li, C. Three-view cotton flower counting through multi-object tracking and RGB-D imagery. Biosyst. Eng. 2024, 246, 233–247. [Google Scholar] [CrossRef]
- Tan, C.; Li, C.; Sun, J.; Song, H. Three-View Cotton Flower Counting through Multi-Object Tracking and Multi-Modal Imaging. In Proceedings of the 2023 ASABE Annual International Meeting, Omaha, NE, USA, 9–12 July 2023; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2023; p. 1. [Google Scholar]
- Xu, R.; Li, C. A review of high-throughput field phenotyping systems: Focusing on ground robots. Plant Phenomics 2022, 2022, 9760269. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, C. Convolutional neural networks for image-based high-throughput plant phenotyping: A review. Plant Phenomics 2020, 2020, 4152816. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.X.; Li, Y.; Wang, R.F.; Hu, P.; Su, W.H. Deep Learning in Multimodal Fusion for Sustainable Plant Care: A Comprehensive Review. Sustainability 2025, 17, 5255. [Google Scholar] [CrossRef]






| Parameters | YOLOv4-tiny | YOLOv5-m | YOLOv7-tiny | YOLOX-tiny | Ours |
|---|---|---|---|---|---|
| mAP/% | 86.40 | 93.19 | 88.50 | 93.49 | 95.16 |
| Precision/% | 97.84 | 99.74 | 99.42 | 97.90 | 98.46 |
| Recall/% | 79.20 | 85.79 | 73.63 | 89.17 | 89.20 |
| F1-Score/% | 88 | 92 | 85 | 93 | 94 |
| Memory usage/M | 5.874 M | 21.056 M | 6.014 M | 5.033 M | 5.055 M |
| Model’s size | 22.40 M | 80.64 M | 23.10 M | 19.40 M | 19.50 M |
| FPS/(f·s−1) | 16.4 | 4.3 | 13.3 | 11.7 | 10.7 |
| Number | Plant Height Measured by the Algorithm (cm) | Actual Plant Height (cm) | Error (cm) |
|---|---|---|---|
| 1 | 21.62 | 21.00 | 0.62 |
| 2 | 20.70 | 21.10 | 0.40 |
| 3 | 21.77 | 22.30 | 0.53 |
| 4 | 26.82 | 26.50 | 0.32 |
| 5 | 18.07 | 18.50 | 0.43 |
| 6 | 20.32 | 21.10 | 0.78 |
| 7 | 20.53 | 20.20 | 0.33 |
| 8 | 22.22 | 22.50 | 0.28 |
| 9 | 22.88 | 22.50 | 0.38 |
| 10 | 19.94 | 20.30 | 0.36 |
| 11 | 20.84 | 21.50 | 0.66 |
| 12 | 22.11 | 21.50 | 0.61 |
| 13 | 22.09 | 22.60 | 0.51 |
| 14 | 19.38 | 20.00 | 0.62 |
| 15 | 24.48 | 24.60 | 0.12 |
| 16 | 20.65 | 21.30 | 0.65 |
| 17 | 21.05 | 21.30 | 0.25 |
| 18 | 22.96 | 22.60 | 0.36 |
| 19 | 30.98 | 31.00 | 0.02 |
| 20 | 21.16 | 21.80 | 0.64 |
| Number | Stem Thickness Measured by Algorithm (mm) | Actual Stem Thickness (mm) | Error (mm) |
|---|---|---|---|
| 1 | 2.97 | 2.98 | 0.01 |
| 2 | 2.61 | 2.72 | 0.11 |
| 3 | 3.06 | 3.03 | 0.03 |
| 4 | 2.88 | 3.01 | 0.13 |
| 5 | 2.70 | 2.73 | 0.03 |
| 6 | 3.38 | 3.35 | 0.03 |
| 7 | 2.97 | 2.83 | 0.14 |
| 8 | 2.88 | 3.08 | 0.20 |
| 9 | 3.12 | 3.15 | 0.03 |
| 10 | 3.38 | 3.30 | 0.08 |
| 11 | 3.06 | 3.23 | 0.17 |
| 12 | 3.64 | 3.68 | 0.04 |
| 13 | 2.97 | 3.11 | 0.14 |
| 14 | 3.24 | 3.33 | 0.09 |
| 15 | 2.90 | 2.92 | 0.02 |
| 16 | 2.82 | 2.80 | 0.02 |
| 17 | 3.00 | 2.93 | 0.07 |
| 18 | 3.10 | 3.02 | 0.08 |
| 19 | 3.16 | 3.10 | 0.06 |
| 20 | 3.00 | 2.95 | 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, Y.; Wang, R.-F.; Zhao, C.-T.; Hu, P.; Wang, H. Research on Obtaining Pepper Phenotypic Parameters Based on Improved YOLOX Algorithm. AgriEngineering 2025, 7, 209. https://doi.org/10.3390/agriengineering7070209
Huo Y, Wang R-F, Zhao C-T, Hu P, Wang H. Research on Obtaining Pepper Phenotypic Parameters Based on Improved YOLOX Algorithm. AgriEngineering. 2025; 7(7):209. https://doi.org/10.3390/agriengineering7070209
Chicago/Turabian StyleHuo, Yukang, Rui-Feng Wang, Chang-Tao Zhao, Pingfan Hu, and Haihua Wang. 2025. "Research on Obtaining Pepper Phenotypic Parameters Based on Improved YOLOX Algorithm" AgriEngineering 7, no. 7: 209. https://doi.org/10.3390/agriengineering7070209
APA StyleHuo, Y., Wang, R.-F., Zhao, C.-T., Hu, P., & Wang, H. (2025). Research on Obtaining Pepper Phenotypic Parameters Based on Improved YOLOX Algorithm. AgriEngineering, 7(7), 209. https://doi.org/10.3390/agriengineering7070209








