Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods
Abstract
:1. Introduction
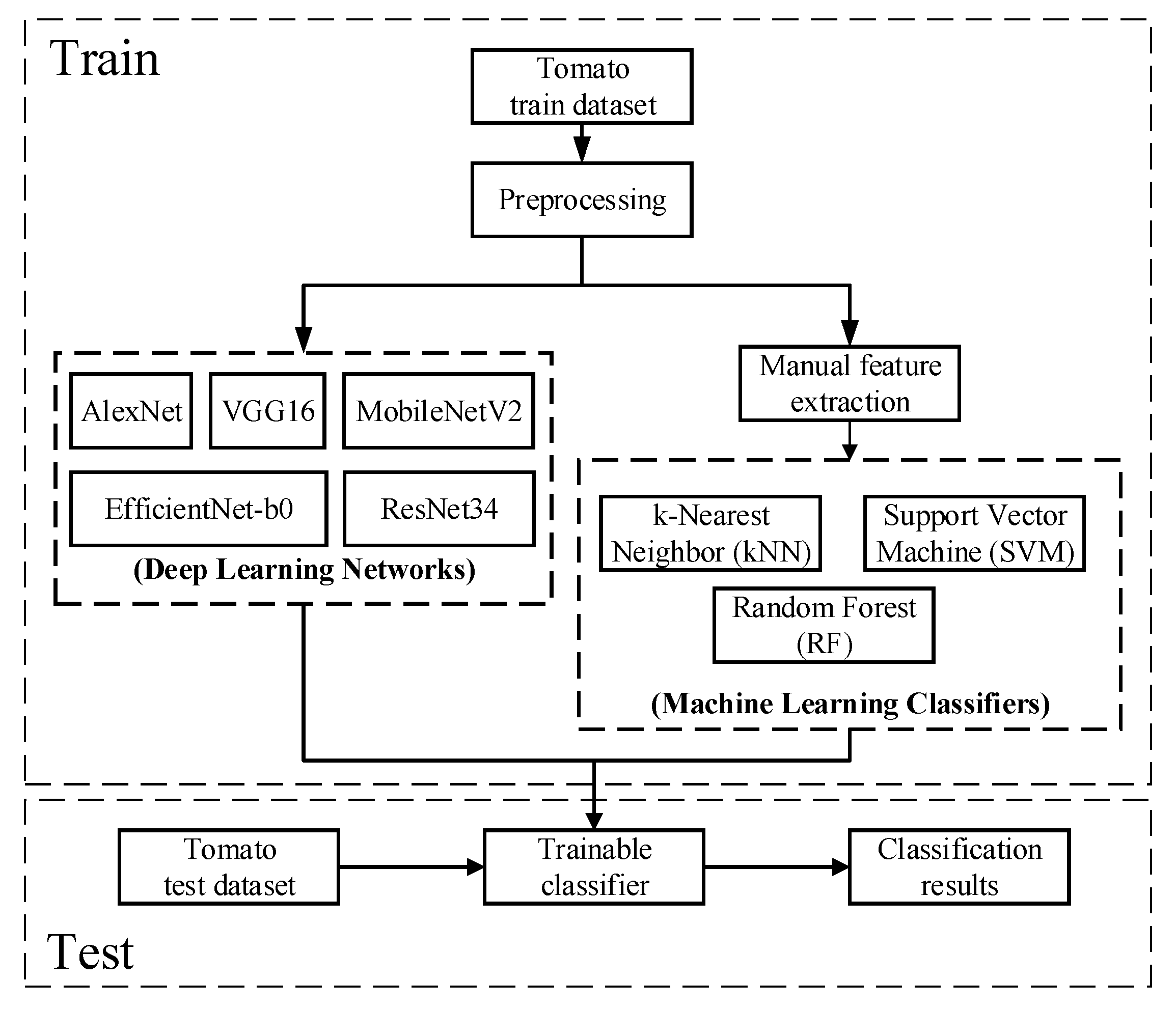
2. Materials and Methods
2.1. Image Acquisition
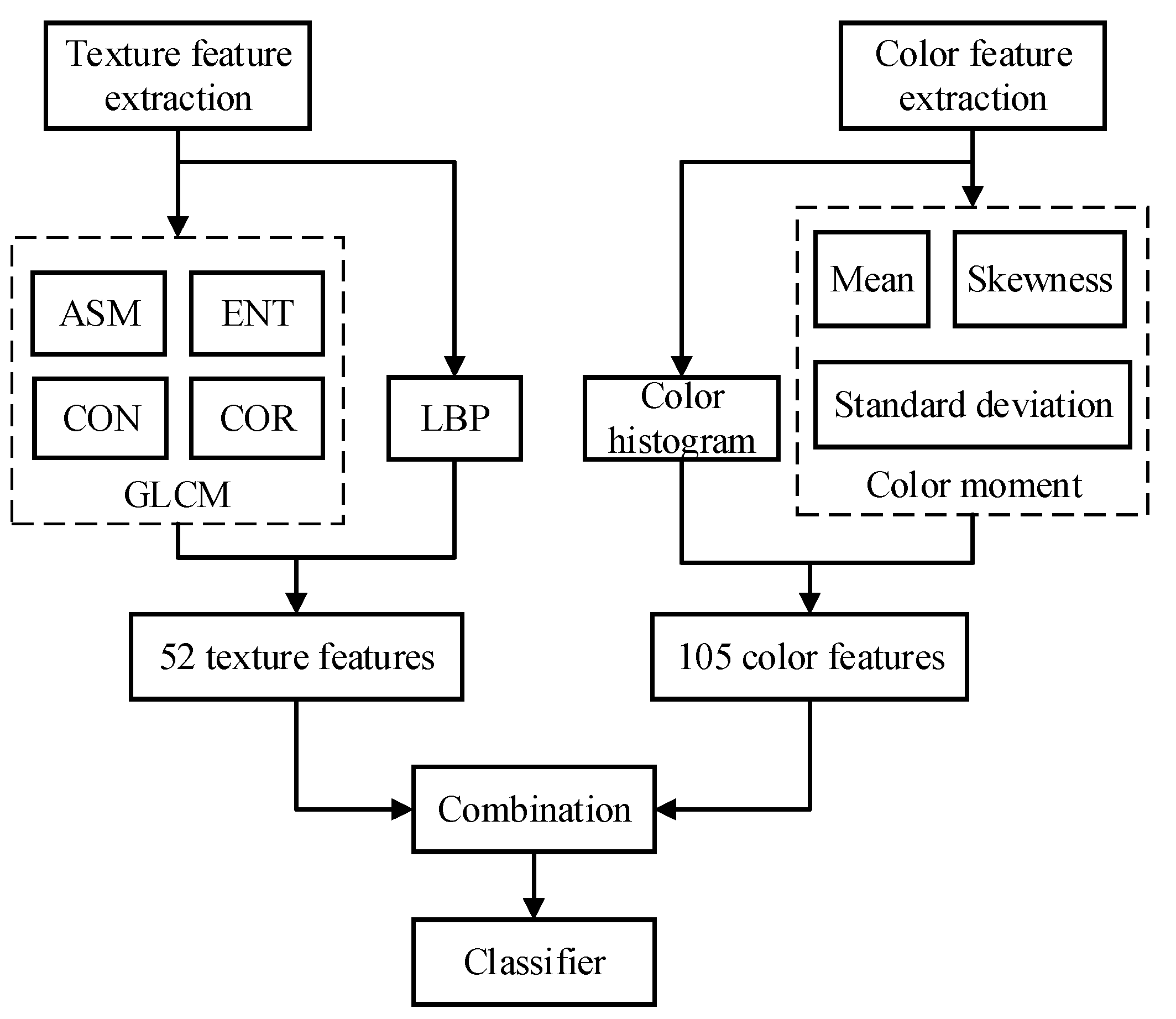
2.2. Texture Features Extraction
2.3. Color Features Extraction
2.4. Machine Learning Classification Methods
2.5. Deep Learning Classification Methods
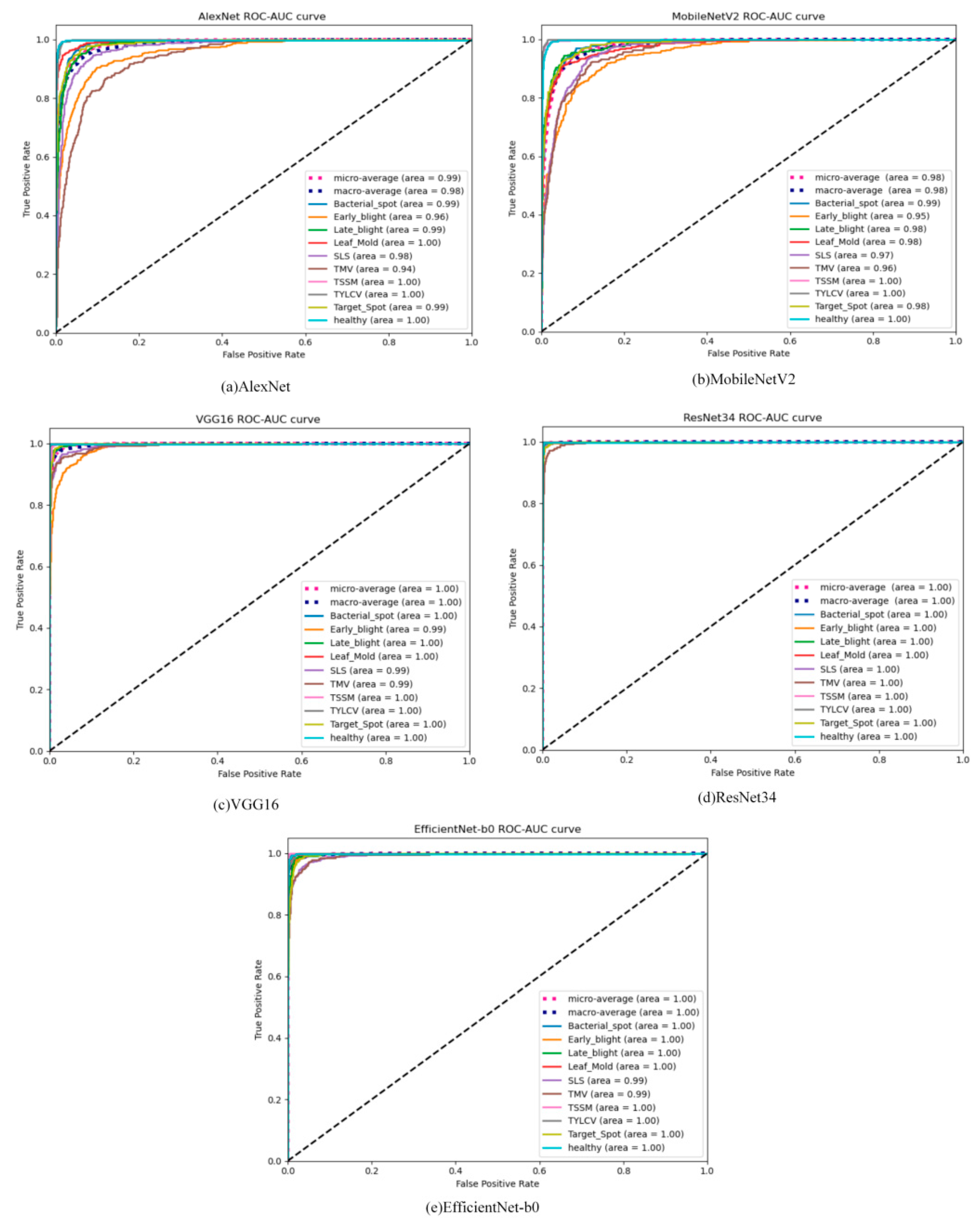
3. Results and Discussion
3.1. Results of Tested ML/DL Algorithms
3.2. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Batuman, O.; Kuo, Y.-W.; Palmieri, M.; Rojas, M.R.; Gilbertson, R.L. Tomato chocolate spot virus, a member of a new torradovirus species that causes a necrosis-associated disease of tomato in Guatemala. Arch. Virol. 2010, 155, 857–869. [Google Scholar] [CrossRef] [Green Version]
- Gleason, M.L. Recent Progress in Understanding and Controlling Bacterial Canker of Tomato in Eastern North America. Plant Dis. 1993, 77, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Xing, J.; Symons, S.; Shahin, M.; Hatcher, D. Detection of sprout damage in Canada Western Red Spring wheat with multiple wavebands using visible/near-infrared hyperspectral imaging. Biosyst. Eng. 2010, 106, 188–194. [Google Scholar] [CrossRef]
- Jones, C.; Jones, J.; Lee, W.S. Diagnosis of bacterial spot of tomato using spectral signatures. Comput. Electron. Agric. 2010, 74, 329–335. [Google Scholar] [CrossRef]
- Ye, H.; Huang, W.; Huang, S.; Cui, B.; Dong, Y.; Guo, A.; Ren, Y.; Jin, Y. Identification of banana fusarium wilt using supervised classification algorithms with UAV-based multi-spectral imagery. Int. J. Agric. Biol. Eng. 2020, 13, 136–142. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, B.; Zhang, X.; Liu, P.; Dong, Y.; Wu, K.; Huang, W. Impact of spectral interval on wavelet features for detecting wheat yellow rust with hyperspectral data. Int. J. Agric. Biol. Eng. 2018, 11, 138–144. [Google Scholar] [CrossRef] [Green Version]
- Vigneau, N.; Ecarnot, M.; Rabatel, G.; Roumet, P. Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in Wheat. Field Crop. Res. 2011, 122, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Stroppiana, D.; Boschetti, M.; Brivio, P.A.; Bocchi, S. Plant nitrogen concentration in paddy rice from field canopy hyperspectral radiometry. Field Crop. Res. 2009, 111, 119–129. [Google Scholar] [CrossRef]
- Zhao, D.; Reddy, K.R.; Kakani, V.G.; Reddy, V. Nitrogen deficiency effects on plant growth, leaf photosynthesis, and hyperspectral reflectance properties of sorghum. Eur. J. Agron. 2005, 22, 391–403. [Google Scholar] [CrossRef]
- Panda, S.S.; Ames, D.P.; Panigrahi, S. Application of Vegetation Indices for Agricultural Crop Yield Prediction Using Neural Network Techniques. Remote Sens. 2010, 2, 673–696. [Google Scholar] [CrossRef] [Green Version]
- Brantley, S.T.; Zinnert, J.C.; Young, D.R. Application of hyperspectral vegetation indices to detect variations in high leaf area index temperate shrub thicket canopies. Remote Sens. Environ. 2011, 115, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral Vegetation Indices and Their Relationships with Agricultural Crop Characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, M.; Gao, Y.; Jiang, H. Using hyperspectral imaging to discriminate yellow leaf curl disease in tomato leaves. Precis. Agric. 2017, 19, 379–394. [Google Scholar] [CrossRef]
- Schubert, T.S.; Rizvi, S.A.; Sun, X.; Gottwald, T.R.; Graham, J.H.; Dixon, W.N. Meeting the challenge of Eradicating Citrus Canker in Florida—Again. Plant Dis. 2001, 85, 340–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarbrick, P.J.; Schulze-Lefert, P.; Scholes, J.D. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 2006, 29, 1061–1076. [Google Scholar] [CrossRef]
- Mojjada, R.K.; Kumar, K.K.; Yadav, A.; Prasad, B.S.V. Detection of plant leaf disease using digital image processing. Mater. Today. 2020. [Google Scholar] [CrossRef]
- Senthilkumar, T.; Jayas, D.S.; White, N.D.; Fields, P.G.; Gräfenhan, T. Detection of fungal infection and Ochratoxin A contamination in stored barley using near-infrared hyperspectral imaging. Biosyst. Eng. 2016, 147, 162–173. [Google Scholar] [CrossRef]
- Lopez-Maestresalas, A.; Keresztes, J.C.; Goodarzi, M.; Arazuri, S.; Jarén, C.; Saeys, W. Non-destructive detection of blackspot in potatoes by Vis-NIR and SWIR hyperspectral imaging. Food Control. 2016, 70, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Karuppiah, K.; Senthilkumar, T.; Jayas, D.; White, N. Detection of fungal infection in five different pulses using near-infrared hyperspectral imaging. J. Stored Prod. Res. 2016, 65, 13–18. [Google Scholar] [CrossRef]
- Jafari, M.; Minaei, S.; Safaie, N.; Torkamani-Azar, F. Early detection and classification of powdery mildew-infected rose leaves using ANFIS based on extracted features of thermal images. Infrared Phys. Technol. 2016, 76, 338–345. [Google Scholar] [CrossRef]
- Lu, J.; Ehsani, R.; Shi, Y.; De Castro, A.I.; Wang, S. Detection of multi-tomato leaf diseases (late blight, target and bacterial spots) in different stages by using a spectral-based sensor. Entific Rep. 2018, 8, 2793. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Liang, L.; Wang, L.; She, J.; Wu, M. Identification of cash crop diseases using automatic image segmentation algorithm and deep learning with expanded dataset. Comput. Electron. Agric. 2020, 177, 105712. [Google Scholar] [CrossRef]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 2020, 105393. [Google Scholar] [CrossRef]
- Sharma, P.; Berwal, Y.P.S.; Ghai, W. Performance analysis of deep learning CNN models for disease detection in plants using image segmentation. Inf. Process. Agric. 2020, 7, 566–574. [Google Scholar] [CrossRef]
- Awan, F.M.; Saleem, Y.; Minerva, R.; Crespi, N. A Comparative Analysis of Machine/Deep Learning Models for Parking Space Availability Prediction. Sensors 2020, 20, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, D.; Salathe, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Radovanovic, D.; Dukanovic, S. Image-Based Plant Disease Detection: A Comparison of Deep Learning and Classical Machine Learning Algorithms. In Proceedings of the 2020 24th International Conference on Information Technology (IT), Zabljak, Montenegro, 18–22 February 2020. [Google Scholar]
- Devassy, B.M.; George, S. Dimensionality reduction and visualisation of hyperspectral ink data using t-SNE. Forensic Sci. Int. 2020, 311, 110194. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathe, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [Green Version]
- Grigorescu, S.E.; Petkov, N.; Kruizinga, P. Comparison of texture features based on Gabor filters. IEEE Trans. Image Process. 2002, 11, 1160–1167. [Google Scholar] [CrossRef] [Green Version]
- Manjunath, B.; Ma, W. Texture features for browsing and retrieval of image data. IEEE Trans. Pattern Anal. Mach. Intell. 1996, 18, 837–842. [Google Scholar] [CrossRef] [Green Version]
- Unser, M.; Eden, M. Multiresolution feature extraction and selection for texture segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 1989, 11, 717–728. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-M.; Chen, Y.-C.; Hsieh, K.-S. Texture features for classification of ultrasonic liver images. IEEE Trans. Med Imaging 1992, 11, 141–152. [Google Scholar] [CrossRef]
- Yang, S.-W.; Lin, C.-S.; Lin, S.-K.; Tseng, Y.-C. Automatic inspection system for defects of printed art tile based on texture feature analysis. Instrum. Sci. Technol. 2013, 42, 59–71. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, N.; Jiang, H. Discriminating Leaves Affected with Tomato Yellow Leaf Curl through Fluorescence Imaging Using Texture and Leaf Vein Features. Trans. Asae Am. Soc. Agric. Eng. 2016, 59, 1507–1515. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Harwood, D. Performance evaluation of texture measures with classification based on Kullback discrimination of distributions. Pattern Recognition—Conference A: Computer Vision and Image Processing. In Proceedings of the 12th IAPR International Conference, Jerusalem, Israel, 9–13 October 1994. [Google Scholar]
- Ojala, T.; Pietik Inen, M.; Harwood, D.A. Comparative Study of Texture Measures with Classification Based on Feature Distri-butions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Ojala, T.; Valkealahti, K.; Oja, E.; Pietikäinen, M. Texture discrimination with multidimensional distributions of signed gray-level differences. Pattern Recognit. 2001, 34, 727–739. [Google Scholar] [CrossRef] [Green Version]
- Pietik Inen, M.; Ojala, T.; Xu, Z. Rotation-invariant texture classification using feature distributions. Pattern Recognit. 2000, 33, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Mäenpää, T.; Pietikäinen, M. Texture analysis with local binary patterns. In Handbook of Pattern Recognition and Computer Vision, 3rd ed.; World Scientific Pub Co Pte Lt: Singapore, 2005; Volume 1, pp. 197–216. [Google Scholar]
- Stricker, M.A.; Orengo, M. Similarity of color images. In Storage and Retrieval for Image and Video Databases III; International Society for Optics and Photonics: Bellingham, WA, USA, 1995; Volume 2420, pp. 381–392. [Google Scholar] [CrossRef]
- Swain, M.J.; Ballard, D.H. Color indexing. Int. J. Comput. Vis. 1991, 7, 11–12. [Google Scholar] [CrossRef]
- Abdullah, N.E.; Rahim, A.A.; Hashim, H.; Kamal, M.M. Classification of Rubber Tree Leaf Diseases Using Multilayer Perceptron Neural Network. In Proceedings of the 2007 5th Student Conference on Research Development, Selangor, Malaysia, 12–11 December 2007. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar]
- Sujatha, R.; Chatterjee, J.M.; Jhanjhi, N.; Brohi, S.N. Performance of deep learning vs machine learning in plant leaf disease detection. Microprocess. Microsyst. 2021, 80, 103615. [Google Scholar] [CrossRef]
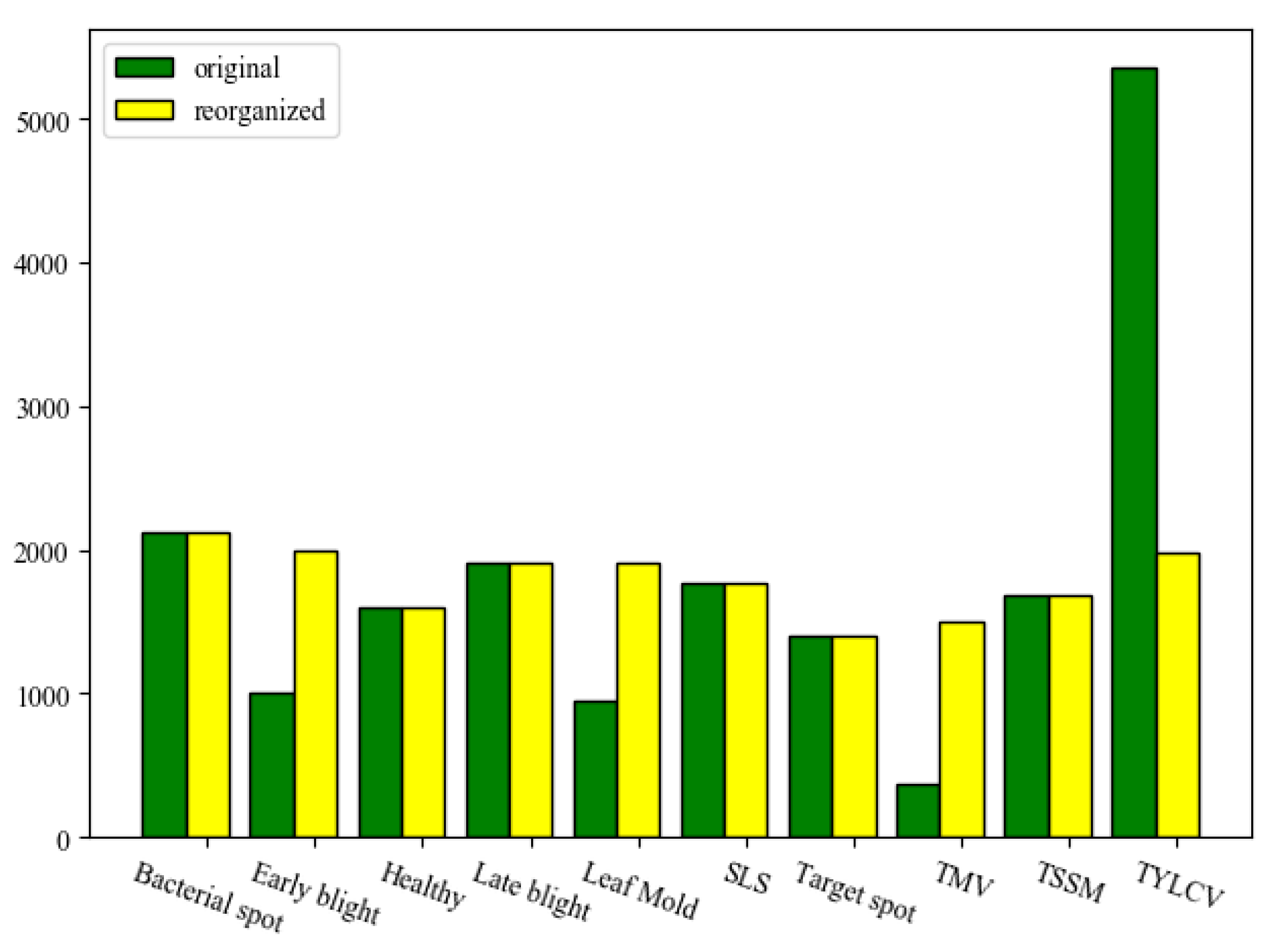
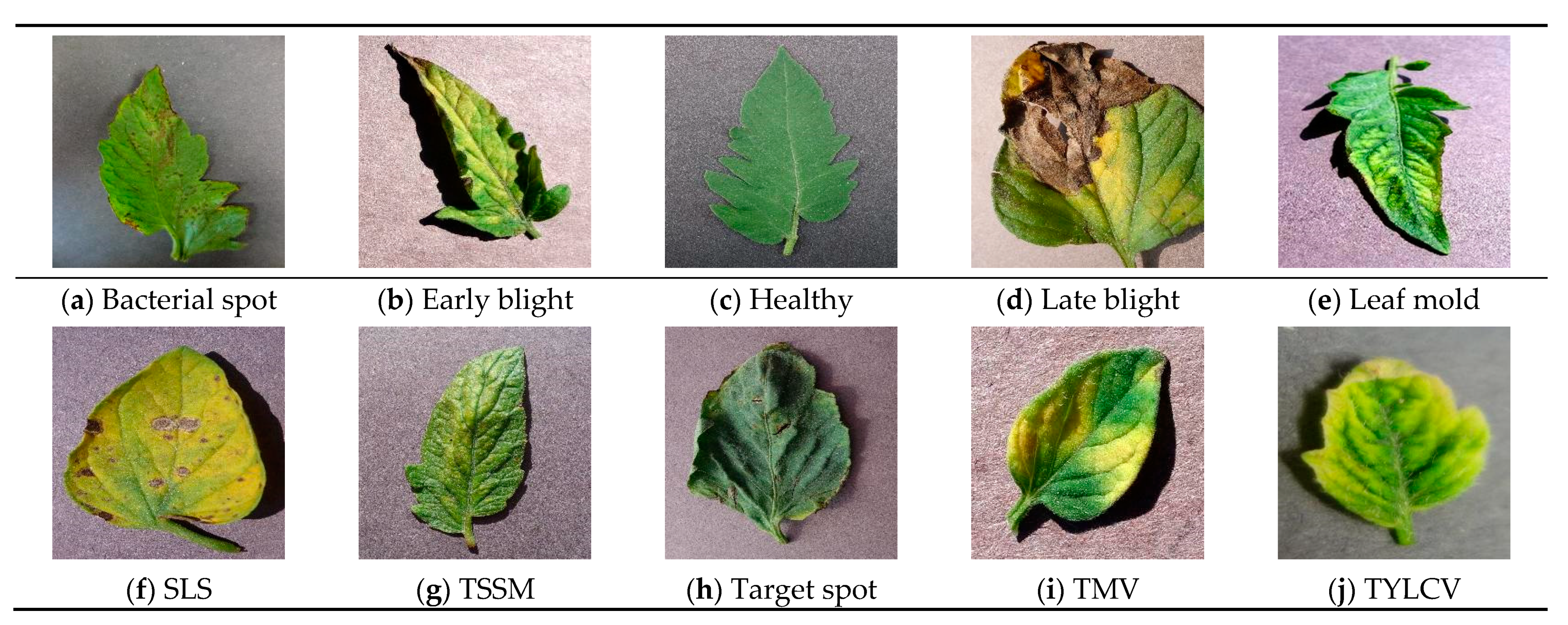



| Disease Common Name | Disease Scientific Name | Image Number |
|---|---|---|
| Bacterial spot | Xanthomonas campestris pv.Vesicatoria | 2127 |
| Early blight | Alternaria solani | 1000 |
| Late blight | Phytophthora infestans | 1909 |
| Leaf mold | Fulvia fulva | 952 |
| Septoria leaf spot (SLS) | Septoria lycopersici | 1771 |
| Two-spotted spider mite (TSSM) | Tetranychus urticae | 1676 |
| Target spot | Corynespora cassiicola | 1404 |
| Tomato mosaic virus (TMV) | Tomato mosaic virus | 373 |
| Tomato yellow leaf curl virus (TYLCV) | Begomovirus (Fam. Geminiviridae) | 5357 |
| Disease Type | Feature Extraction Method | ||||||
|---|---|---|---|---|---|---|---|
| LBP | GLCM | LBP+GLCM | COLOR | COLOR+LBP | COLOR+GLCM | ALL | |
| Bacterial spot | (57,75) | (67,87) | (71,92) | (83,79) | (85,85) | (85,97) | (85,90) |
| Early blight | (52,41) | (66,57) | (73,58) | (83,64) | (81,72) | (85,76) | (81,78) |
| Late blight | (59,71) | (62,77) | (78,86) | (68,92) | (80,93) | (90,97) | (90,95) |
| Leaf mold | (64,44) | (66,58) | (70,61) | (73,43) | (78,55) | (78,69) | (82,65) |
| SLS | (63,65) | (60,43) | (69,65) | (68,75) | (76,82) | (81,85) | (75,84) |
| TMV | (61,49) | (56,49) | (70,59) | (77,66) | (83,71) | (87,82) | (85,78) |
| TSSM | (51,62) | (57,72) | (66,79) | (63,76) | (75,84) | (80,88) | (79,86) |
| TYLCV | (43,43) | (49,58) | (55,62) | (63,67) | (67,70) | (78,80) | (74,72) |
| Target spot | (75,83) | (75,81) | (81,86) | (78,99) | (79,100) | (93,95) | (89,98) |
| Healthy | (76,73) | (67,53) | (85,73) | (82,87) | (88,90) | (99,89) | (91,89) |
| Average | (60,61) | (63,63) | (72,72) | (74,75) | (79,80) | (86,86) | (83,83) |
| Methods | Evaluation Metrics | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | |
| kNN | 82.1% | 82.1% | 82.5% | 82.0% |
| SVM | 91.0% | 90.9% | 91.0% | 91.0% |
| Random forest | 82.7% | 82.6% | 83.0% | 82.7% |
| AlexNet | 92.7% | 92.5% | 92.6% | 92.4% |
| VGG16 | 98.9% | 98.6% | 98.5% | 98.5% |
| ResNet34 | 99.7% | 99.6% | 99.7% | 99.7% |
| EfficientNet-b0 | 98.9% | 98.9% | 98.9% | 98.8% |
| MobileNetV2 | 91.2% | 91.3% | 91.3% | 91.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, L.; Lu, J.; Jiang, H. Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods. AgriEngineering 2021, 3, 542-558. https://doi.org/10.3390/agriengineering3030035
Tan L, Lu J, Jiang H. Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods. AgriEngineering. 2021; 3(3):542-558. https://doi.org/10.3390/agriengineering3030035
Chicago/Turabian StyleTan, Lijuan, Jinzhu Lu, and Huanyu Jiang. 2021. "Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods" AgriEngineering 3, no. 3: 542-558. https://doi.org/10.3390/agriengineering3030035
APA StyleTan, L., Lu, J., & Jiang, H. (2021). Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods. AgriEngineering, 3(3), 542-558. https://doi.org/10.3390/agriengineering3030035






