Evaluation of Structured, Semi-Structured, and Free-Text Electronic Health Record Data to Classify Hepatitis C Virus (HCV) Infection
Abstract
1. Introduction
2. Methods
Data Source and Study Design
3. Methodology of Risk Prediction Model
3.1. Variables
3.2. Structured Data Elements
3.3. Semi-Structured Data Elements
3.4. Free-Text Data Elements
4. Model Development and Validation
5. Results
6. Model Results
7. Prevalence of Substance and Opioid Use Amongst Predictors
8. Comparison of Structured and Unstructured Extraction
9. Discussion
9.1. Utility of Structured Fields
9.2. Free-Text Concepts
9.3. HCV and Opioid Use
9.4. Patient Care
10. Limitations
11. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Combating Hepatitis B and C to Reach Elimination by 2030: Advocacy Brief. 2016. Available online: https://apps.who.int/iris/handle/10665/206453 (accessed on 5 May 2022).
- HCV in Pregnancy. 2018. Available online: https://www.hcvguidelines.org/unique-populations/pregnancy (accessed on 5 May 2022).
- Indian Health Service Highlights Initiative to Eliminate Hepatitis C and HIV/AIDS in Indian Country during National Native HIV/AIDS Awareness Day|2019 Press Releases. 2019. Available online: https://www.ihs.gov/newsroom/pressreleases/2019pressreleases/indian-health-service-highlights-initiative-to-eliminate-hepatitis-c-and-hiv-aids-in-indian-country-during-national-native-hiv-aids-awareness-day/ (accessed on 5 May 2022).
- Denniston, M.M.; Klevens, R.M.; McQuillan, G.M.; Jiles, R.B. Awareness of infection, knowledge of hepatitis C, and medical follow-up among individuals testing positive for hepatitis C: National Health and Nutrition Examination Survey 2001–2008. Hepatology 2012, 55, 1652–1661. [Google Scholar] [CrossRef] [PubMed]
- Waruingi, W.; Mhanna, M.; Kumar, D.; Abughali, N. Hepatitis C virus universal screening versus risk based selective screening during pregnancy. J. Neonatal Perinat. Med. 2015, 8, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Chaillon, A.; Rand, E.B.; Reau, N.; Martin, N.K. Cost-effectiveness of Universal Hepatitis C Virus Screening of Pregnant Women in the United States. Clin. Infect. Dis. 2019, 69, 1888–1895. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.D.; Morgan, R.L.; Beckett, G.A.; Falck-Ytter, Y.; Holtzman, D.; Teo, C.G.; Ward, J.W. Recommendations for the Identification of Chronic Hepatitis C Virus Infection Among Persons Born During 1945–1965. MMWR 2012, 61, 1–32. [Google Scholar] [PubMed]
- Barocas, J.A.; Tasillo, A.; Eftekhari Yazdi, G.; Wang, J.; Vellozzi, C.; Hariri, S.; Isenhour, C.; Randall, L.; Ward, J.W.; Mermin, J.; et al. Population-level outcomes and cost-effectiveness of expanding the recommendation for age-based hepatitis C testing in the United States. Clin. Infect. Dis. 2018, 67, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, B.; Perumalswami, P.V.; Mageras, A.; Miller, M.; Harty, A.; Ma, N.; Bowman, C.A.; Collado, F.; Jeon, J.; Paulino, L.; et al. A Digital Case-Finding Algorithm for Diagnosed but Untreated Hepatitis C: A Tool for Increasing Linkage to Treatment and Cure. Hepatology 2021, 74, 2974–2987. [Google Scholar] [CrossRef] [PubMed]
- Nandipati, S.C.; XinYing, C.; Wah, K.K. Hepatitis C virus (HCV) prediction by machine learning techniques. Appl. Model. Simul. 2020, 4, 89–100. [Google Scholar]
- Friedman, C.; Johnson, S.B. Natural Language and Text Processing in Biomedicine. In Biomedical Informatics: Computer Applications in Health Care and Biomedicine; Springer: Berlin/Heidelberg, Germany, 2006; pp. 312–343. [Google Scholar]
- Spyns, P. Natural Language Processing in Medicine: An Overview. Methods Inf. Med. 1996, 35, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Chapman, W.W.; Nadkarni, P.M.; Hirschman, L.; D’Avolio, L.W.; Savova, G.K.; Uzuner, O. Overcoming barriers to NLP for clinical text: The role of shared tasks and the need for additional creative solutions. J. Am. Med. Inform. Assoc. JAMIA 2011, 18, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Fong, A.; Adams, K.T.; Gaunt, M.; Howe, J.L.; Kellogg, K.M.; Ratwani, R.M. Identifying Health Information Technology Related Safety Event Reports from Patient Safety Event Report Databases. J. Biomed. Inform. 2018, 86, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Khanbhai, M.; Anyadi, P.; Symons, J.; Flott, K.; Darzi, A.; Mayer, E. Applying natural language processing and machine learning techniques to patient experience feedback: A systematic review. BMJ Health Care Inform. 2021, 28, e100262. [Google Scholar] [CrossRef] [PubMed]
- Moons, K.G.M.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.A.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.L.; Hurley, L.B.; Krakower, D.S.; Alexeeff, S.; Silverberg, M.J.; Volk, J.E. Use of electronic health record data and machine learning to identify candidates for HIV pre-exposure prophylaxis: A modelling study. Lancet HIV 2019, 6, e688–e695. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13 August 2016. [Google Scholar]
- Pfob, A.; Sidey-Gibbons, C.; Lee, H.-B.; Tasoulis, M.K.; Koelbel, V.; Golatta, M.; Rauch, G.M.; Smith, B.D.; Valero, V.; Han, W.; et al. Identification of breast cancer patients with pathologic complete response in the breast after neoadjuvant systemic treatment by an intelligent vacuum-assisted biopsy. Eur. J. Cancer 2021, 143, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Tabaie, A.; Orenstein, E.W.; Nemati, S.; Basu, R.K.; Kandaswamy, S.; Clifford, G.D.; Kamaleswaran, R. Predicting presumed serious infection among hospitalized children on central venous lines with machine learning. Comput. Biol. Med. 2021, 132, 104289. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Kumar, V.; Ross Quinlan, J.; Ghosh, J.; Yang, Q.; Motoda, H.; McLachlan, G.J.; Ng, A.; Liu, B.; Yu, P.S.; et al. Top 10 algorithms in data mining. Knowl. Inf. Syst. 2008, 14, 1–37. [Google Scholar] [CrossRef]
- Quinlan, J.R. C4.5: Program for Machine Learning; Elsevier: Amsterdam, The Netherlands, 1993. [Google Scholar]
- Observational Health Data Sciences and Informatics. Standardized Data: The OMOP Common Data Model. 2022. Available online: https://www.ohdsi.org/data-standardization/ (accessed on 2 February 2022).
- Armstrong, G.L.; Wasley, A.; Simard, E.P.; McQuillan, G.M.; Kuhnert, W.L.; Alter, M.J. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 2006, 144, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.; Alpert, A.; Pacula, R.L. A transitioning epidemic: How the opioid crisis is driving the rise in hepatitis C. Health Aff. 2019, 38, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Smart, A.; Geboy, A.; Basch, P.; Nichols, W.; Zeymo, A.; Perez, I.; Hafeez, M.; Fleisher, I.; Fernandez, S.; Fishbein, D. Identification of risk factors for testing of hepatitis C in non-birth cohort patients: Is universal screening necessary? J. Addict. Med. 2021, 15, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Hack, B.; Timalsina, U.; Tefera, E.; Wilkerson, B.; Paku, E.; Fernandez, S.; Fishbein, D. Oral prescription opioids as a high-risk indicator for hepatitis C infection: Another step toward HCV elimination. J. Prim. Care Community Health 2021, 12, 21501327211034379. [Google Scholar] [CrossRef] [PubMed]
| Variable (n) | ICD10 Codes and Other Variable Descriptions with Examples |
|---|---|
| Structured Data Elements (47) | |
| Sexually transmitted infections (14) | A53.9, A54.6, A55, A56.3, A57, A58, A60.1, B00, Z11.4, Z11.3, Z20.6, Z11.59, Z20.2, Z72.5 (Diagnosis) |
| Viral hepatitis and HIV (7) | B17.1, B18.1, B18.2, B19.1, B19.2, Z20.5, B20 (Diagnosis) |
| Substance use disorders (6) | F11, F14, F19.2, K62.89, Z79.899, Z71.5 (Diagnosis) |
| MEDS_opioid | Opioid medication (Medication) |
| Liver disease and laboratory abnormalities (12) | K70, K71, K72, K73, K74, K75, K76, K77, R74.01, R94.5, W46.0, Z01.812 (Diagnosis) |
| Other medical co-morbidities (3) | N18.6, Z99.2, Z94 (Diagnosis) |
| Other factors influencing health status (4) | Z65.1, Z72.89, Z77.21, Z86.59 (Diagnosis) |
| Semi-structured and free-text data elements (7) | |
| TEXT_Substance | Patient with history of substance abuse or use including methamphetamine, cocaine, heroin *look up slang* and excludes tobacco, nicotine, and marijuana Included example “female with history of COPD, depression, substance abuse” Excluded example “Substance Use: Denies” |
| TEXT_Incarceration | Patient have been incarcerated Included example: “Incarcerated 2003 for joyriding” Excluded example: “father being incarcerated” |
| TEXT_Piercing | Presence of piercing on patient Included example: “Piercing or Tattoo: Professional Piercing’ Excluded example: “Piercing or Tattoo: No’ |
| TEXT_Tattoo | Presence of tattoo on patient Included example: “Piercing or Tattoo: Professional Tattoo’ Excluded example: “Piercing or Tattoo: No’ |
| TEXT_Transfusion | Patient with history of transfusions and solid organ transplants prior to 1992 Included example: “last transfusion on” Excluded example: “did not require transfusion” |
| TEXT_Needlestick | Patient with history of needlestick injury Included example: “sustained a needlestick to right toe” |
| TEXT_STI | Patient with history of sexual transmitted infection through homosexual or bisexual behaviors including syphilis, chlamydia, and gonorrhea Included example “high risk homosexual behavior” |
| Demographics (included) (4): Sex, age, race, ethnicity | |
| Newly Identified HCV Infection Status. No. (%) | ||||
|---|---|---|---|---|
| Characteristic | All (N = 3564) | Newly Identified HCV Infection (n = 487) | No Newly Identified HCV Infection (n = 3077) | p-Value |
| Sex | <0.001 | |||
| Female | 2436 (68.4) | 238 (48.9) | 2198 (71.4) | |
| Male | 1128 (31.6) | 249 (51.1) | 879 (28.6) | |
| Race | <0.001 | |||
| Black | 1617 (45.4) | 256 (52.6) | 1361 (44.2) | |
| White | 1453 (40.8) | 203 (41.7) | 1250 (40.6) | |
| Other | 494 (13.8) | 28 (5.7) | 466 (15.2) | |
| Ethnicity | 0.002 | |||
| Hispanic | 86 (2.4) | 10 (2.1) | 76 (2.5) | |
| Non-Hispanic | 3241 (90.9) | 462 (94.9) | 2779 (90.3) | |
| Other | 237 (6.6) | 15 (3.1) | 222 (7.2) | |
| Age | 45.3 (16.6 SD) | 52.1 (14.5 SD) | 44.3 (16.6 SD) | <0.001 |
| History of HCV prior to 2020 | <0.001 | |||
| Yes | 349 (9.8) | 243 (49.9) | 107 (3.4) | |
| No | 3215 (90.2) | 244 (50.1) | 2970 (96.6) | |
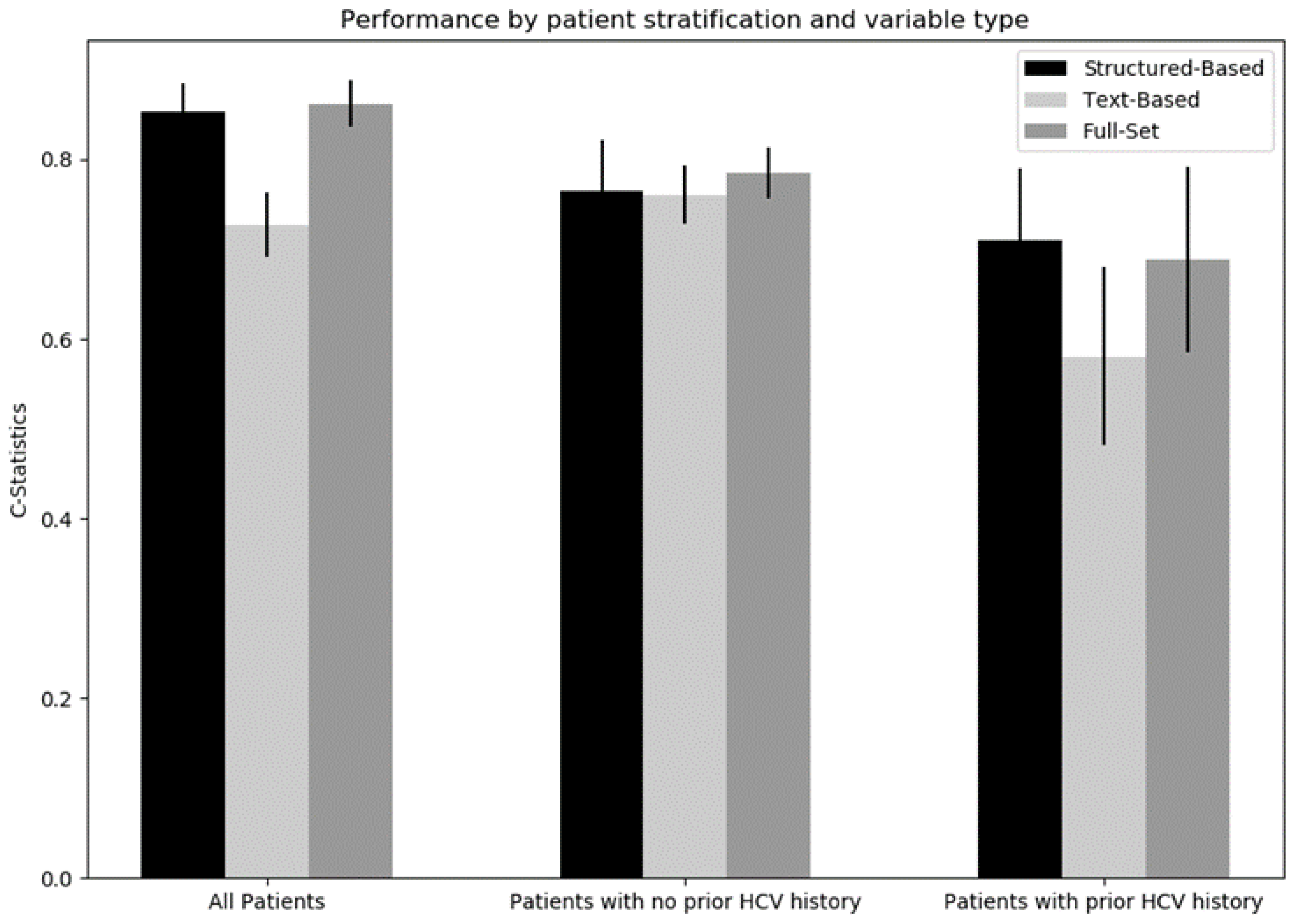
| Structured-Based | Text-Based | Full-Set | |
|---|---|---|---|
| All patients | 0.856 (0.837, 0.876) | 0.725 (0.702, 0.749) | 0.864 (0.845, 0.883) |
| No prior history of HCV | 0.774 (0.735, 0.813) | 0.759 (0.736, 0.781) | 0.780 (0.743, 0.817) |
| Prior history of HCV | 0.700 (0.657, 0.743) | 0.548 (0.478, 0.618) | 0.677 (0.622, 0.732) |
| Average | 0.777 (0.744, 0.810) | 0.677 (0.631, 0.723) | 0.774 (0.735, 0.813) |
| Predictor | Predictor Description | All (n = 3564) | No HCV Infection History (n = 3215) | HCV Infection History (n = 349) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | New HCV Infection (n = 487) | No new HCV Infection (n = 3077) | Importance | Ratio (0.16) | New HCV Infection (n = 244) | No new HCV Infection (n = 2970) | Importance | Ratio (0.08) | New HCV Infection (n = 243) | No New HCV Infection (n = 107) | Importance | Ratio (2.27) | ||
| F11 | Opioid related disorders | 66 | 52 | 14 | 12.471 | 3.71 | 15 | 11 | 11.809 | 1.36 | 37 | 3 | 0 | 12.33 |
| N18.6 | End stage renal disease | 105 | 33 | 72 | 5.318 | 0.46 | 24 | 49 | 6.233 | 0.49 | 9 | 23 | 1.649 | 0.39 |
| Text_Substance | Patient with a history of substance use or abuse | 327 | 146 | 181 | 2.854 | 0.81 | 44 | 146 | 4.492 | 0.30 | 102 | 35 | 0.561 | 2.91 |
| Text_Piercing | Presence of a piercing on patient | 103 | 13 | 90 | 0.492 | 0.14 | 6 | 87 | 2.789 | 0.07 | 7 | 3 | 0.052 | 2.33 |
| F14 | Cocaine related disorders | 16 | 11 | 5 | 2.645 | 2.20 | 2 | 3 | 1.975 | 0.67 | 9 | 2 | 0.59 | 4.50 |
| Z11.3 | Encounter for screening for infections with a predominantly sexual mode of transmission | 454 | 29 | 425 | 1.25 | 0.07 | 7 | 420 | 1.647 | 0.02 | 22 | 5 | 1.088 | 4.40 |
| Meds_Opioid | Opioid medication | 467 | 120 | 347 | 2.295 | 0.35 | 42 | 323 | 1.455 | 0.13 | 78 | 24 | 0.13 | 3.25 |
| Text_STI | Patient with a history of sexual transmitted infection | 1150 | 111 | 1039 | 1.448 | 0.11 | 37 | 1008 | 0.93 | 0.04 | 74 | 31 | 0.411 | 2.39 |
| R94.5 | Abnormal results of liver function studies | 55 | 13 | 42 | 0 | 0.31 | 4 | 36 | 0.721 | 0.11 | 9 | 6 | 0 | 1.50 |
| Z94 | Transplanted organ and tissue status | 84 | 17 | 67 | 15.285 | 0.25 | 8 | 31 | 0.632 | 0.26 | 9 | 36 | 26.074 | 0.25 |
| Z11.59 | Encounter for screening for other viral diseases | 190 | 28 | 162 | 2.029 | 0.17 | 9 | 160 | 0.5 | 0.06 | 19 | 2 | 0 | 9.50 |
| K74 | Fibrosis and cirrhosis of liver | 72 | 34 | 38 | 1.416 | 0.89 | 4 | 16 | 0.474 | 0.25 | 30 | 22 | 2.348 | 1.36 |
| K76 | Other diseases of liver | 93 | 15 | 78 | 2.805 | 0.19 | 0 | 64 | 0 | 0.00 | 15 | 14 | 3.514 | 1.07 |
| Text_transfusion | Patient with a history of transfusions and solid organ transplants prior to 1992 | 622 | 132 | 490 | 1.015 | 0.27 | 39 | 433 | 0 | 0.09 | 93 | 57 | 2.26 | 1.63 |
| Text_tattoo | Presence of a tattoo on patient | 92 | 14 | 78 | 0.836 | 0.18 | 5 | 74 | 0 | 0.07 | 9 | 4 | 0.863 | 2.25 |
| Text Predictor (Associated ICD10) | Total Patients Identified | Identified Only from Free-Text (%) | Identified Only from ICD10 (%) | Identified from Both (%) |
|---|---|---|---|---|
| TEXT_Incarceration (Z65.1) | 107 | 107 (100) | 0 | 0 |
| TEXT_Needlestick (W46) | 11 | 10 (91) | 0 | 1 (9) |
| TEXT_Substance (Z71.5, F11, F14, F19.2) | 352 | 276 (78) | 25 (7) | 51 (14) |
| TEXT_STI (A53.9, A54.6, A55, A56.3, A57, A58, A60.1, B00, B17.1, B18.1, B18.2, B19.1, B19.2, B20, K62.89) | 1422 | 967 (68) | 272 (19) | 183 (13) |
| TEXT_Piercing (Z41.3) | 103 | 103 (100) | 0 | 0 |
| TEXT_Tattoo (L81.8) | 92 | 92 (100) | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fong, A.; Hughes, J.; Gundapenini, S.; Hack, B.; Barkhordar, M.; Huang, S.S.; Visconti, A.; Fernandez, S.; Fishbein, D. Evaluation of Structured, Semi-Structured, and Free-Text Electronic Health Record Data to Classify Hepatitis C Virus (HCV) Infection. Gastrointest. Disord. 2023, 5, 115-126. https://doi.org/10.3390/gidisord5020012
Fong A, Hughes J, Gundapenini S, Hack B, Barkhordar M, Huang SS, Visconti A, Fernandez S, Fishbein D. Evaluation of Structured, Semi-Structured, and Free-Text Electronic Health Record Data to Classify Hepatitis C Virus (HCV) Infection. Gastrointestinal Disorders. 2023; 5(2):115-126. https://doi.org/10.3390/gidisord5020012
Chicago/Turabian StyleFong, Allan, Justin Hughes, Sravya Gundapenini, Benjamin Hack, Mahdi Barkhordar, Sean Shenghsiu Huang, Adam Visconti, Stephen Fernandez, and Dawn Fishbein. 2023. "Evaluation of Structured, Semi-Structured, and Free-Text Electronic Health Record Data to Classify Hepatitis C Virus (HCV) Infection" Gastrointestinal Disorders 5, no. 2: 115-126. https://doi.org/10.3390/gidisord5020012
APA StyleFong, A., Hughes, J., Gundapenini, S., Hack, B., Barkhordar, M., Huang, S. S., Visconti, A., Fernandez, S., & Fishbein, D. (2023). Evaluation of Structured, Semi-Structured, and Free-Text Electronic Health Record Data to Classify Hepatitis C Virus (HCV) Infection. Gastrointestinal Disorders, 5(2), 115-126. https://doi.org/10.3390/gidisord5020012







